International Journal of Scientific & Engineering Research, Volume 4, Issue 8, August-2013 1312
ISSN 2229-5518
Aman Chandra Kaushik
Department of Bioinformatics, University Institute of Engineering & Technology
Chhatrapati Shahu Ji Maharaj University, Kanpur-208024, Uttar Pradesh, India
Email: amanbioinfo@gmail.com
Phone: +91-8924003818
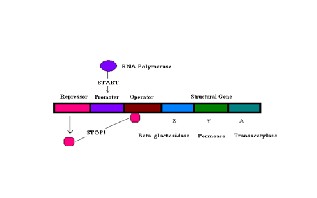
In one circuit described in the paper, one DNA sequences have three genes called Repressor, Operator (terminators) are interposed between the promoter (DNA Polymerase) and the output Proteins (Beta glactosidase, permease, Transacetylase in this case). Each of these terminators inhibits the transcription and Translation of the output gene and can be flipped by a different Promoter enzyme (DNA Polymerase), making the terminator inactive.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 8, August-2013 1313
ISSN 2229-5518
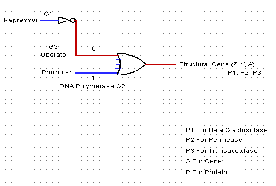
Language and circuits are designed in
Logisim
The underlying molecular automaton is a Boolean network of logic gates that are made of allosterically modulated deoxyribozymes, and input and output are given by oligonucleotides. The logic gates are based on molecular beacons introduced by S. Tyagi and F. Kramer (1996), which are oligonucleotide probes able to report the presence of specific DNA in homogenous solutions. Molecular beacons are hairpin-shaped molecules with a quencher (R) at the 3’ end and a fluorosphore (F) at the 5’ end. These molecules are non- fluorescent as the loop keeps the quencher close to the fluorophore. However, when the oligonucleotide probe sequence in the loop hybridizes to a target DNA molecule forming a rigid double helix, the quencher is separated from the fluorophore restoring fluorescence. Deoxyribozymes are nucleic acid catalysts made of deoxyribonucleic acid. Examples of deoxyribozymes are phosphodiesterases, which cleave other oligonucleotides with shorter products as output, and ligases, which combine two oligonucleotides into a larger product. The molecular gates are based on phosphodiesterase E6, which cleaves an input fluorogenic oligonucleotide upon binding and releases the cleft oligonucleotides as output. This produces an increase in fluorescence by fluorescein (F) as the quencher (R) is separated.
The YES gate is comprised of a deoxyribozyme
E6 module and a hairpin module complementary to input oligonucleotide x. If the input oligonucleotide hybridizes to the hairpin module, the gate enters the active state, increasing fluorescence.
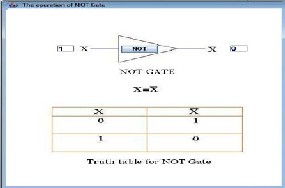
The NOT gate is constructed by extending the non-conserved loop of the E6 core with a hairpin structure that is complementary to input oligonucleotide x. If the input oligonucleotide hybridizes to the hairpin module, the gate enters the inactive state not increasing fluorescence.
The AND gate is obtained from the E6 core module by extending both ends with hairpin modules, one complementary to input oligonucleotide x and the other complementary to input oligonucleotide y.
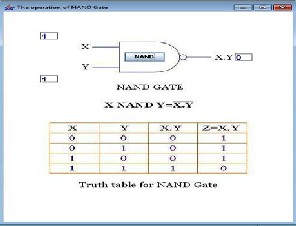
The AND-NOT gate realizes the Boolean function (x, y) _→ xy and is derived from the E6 core module by extending one end with a hairpin module.
Using this design strategy, the researchers can create all two or more input logic gates and implement sequential logic systems for Life Science. It’s really easy to swap things in and out,” says Lu, who is also a member of MIT’s Synthetic Biology Center. “If you start off with a standard parts library, you can use a one- step reaction to assemble any kind of function that you want simply input 1 or 0 and get output according to circuits.”
.
1. Such operon logisim circuits could also be used to create a type of circuit known as a digital-to-analog converter. This kind of circuit takes digital inputs — for example, the presence or absence of single chemicals or molecular pathway— and converts them to an analog output, which can be a range of values, such as continuous levels of gene expression.
2. That type of operon circuit could offer better control over the production of cells or organs or whole body of the System generate biofuels, drugs or other useful compounds. Instead of creating circuits that are always on, or using promoters that need continuous inputs to control their output levels, scientists could transiently program the circuit to produce at a certain level. The cells and their progeny would always remember that level, without needing any more information it is user friendly software put 0 or 1 and get results.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 8, August-2013 1314
ISSN 2229-5518
3. Used as environmental sensors, such circuits could also provide very
precise long-term memory. Java provide JVM (Java Virtual Machine), it provide portable environment for operon logistic circuits.
4. This platform could also allow scientists to more accurately control the fate of stem cells as well as germinal cells they develop into other cell types. Any bio molecular and chemical pathway type of task you perform within a time.
5. Logic and memory are essential functions of circuits that generate complex, state-dependent responses. Here we describe a strategy for efficiently assembling synthetic genetic circuits that use Operon System to implement Boolean logic functions with stable DNA-encoded memory of events. Application of this strategy allowed us to create all two- input Boolean logic functions in living Bacteria cells without requiring cascades comprising multiple logic gates like alignment of two sequences in term of Bioinformatics.
Logisim
JAVA


IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 8, August-2013 1315
ISSN 2229-5518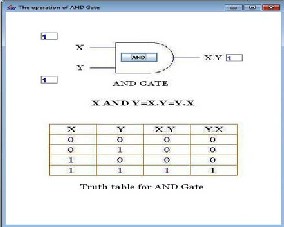
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 8, August-2013 1316
ISSN 2229-5518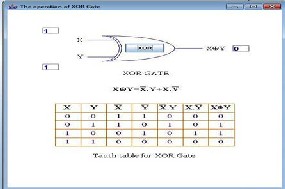
[1] Gheorghe Paun, Computing with Bio- Molecules (Theory and Ex- periments), Springer- Verlag Singapore Pte. Ltd, May 1998.
[2] Leonard M. Adleman, Computing with DNA, Scientific American, August 1998.
[3] | Masahito | Yamamoto, |
Nobuo Toshikazu | Matsuura, Shiba, Yumi |
Kawazoe and Azuma Ohuchi, “Solution of Shortest Path Proble m by Concentration Control,” DNA based computers 5, DIMACS series in Discrete Mathe matics and Theoretical Computer Volume
54, 2000, 101-110.
[4] Gheorghe Paun, Rozenberg and Salomaa, DNA computing, New computing paradigms, Springer-Verlag, Berlin,1998.
[5] L.M Adleman, “Molecular
computation of solutions to combina- torial problems,” Science, 266,
1994, 1021-1024.
[6] Martyn Amos, Gheorghe Paun, Grzezorg, Rozenberg and Arto Salomaa, “Topics in the theory of DNA computing,” Theoretical computer science, 287,
2000, 3-38.
[7] Yachun Liu, Jin Xu, Linqiang and Shiying Wang, “DNA solution to the Graph-Coloring Problem,” J. Chem. Inf, Comput. Sci. 42,
2002, 524-528.
[8] Lloyd Smith, H a ll and Mark, “ Practical feature s u b se t se le c t io n for machine learning,” In Proc Australian C o m p ut e r S c i e n ce Con- ference, Perth, Australia, 1998 , 181-191.
[9] L. Adleman, P.
Rothe mund, S. Roweis and E. Winfree, “On Apply- ing Molecular Computation To The Data Encryption Standard,”
2nd DIMACS workshop on DNA
based computers, Princeton,
1996, 28-48.
[10] Gupta, GauraV, Mehra, Nipun and ChakraVerty, DNA Comput- ing, The Indian Programmer, June
12, 2001.
[11] V. Shekhar, Ch.V.Ramana Murthy, P.V. Gopalacharyulu and G.P.Ra jasekhar, DNA solution to the shortest path problem, ICADS-2002, Dec22-24, Dept of Mathe matics, IIT Kharagpur.
IJSER © 2013 http://www.ijser.org