International Journal of Scientific & Engineering Research Volume 3, Issue 7, June-2012 1
ISSN 2229-5518
1 INSILICO DESIGNING AND DEVELOPMENT OF VACCINE FOR V.CHOLERAE O139 IN
2 CHOLERA DISEASE
3
5 1PES Institute of Technology, Shimoga -580031, India
6 1Research work is carried out at DNA LABS INDIA, Hyderabad, India
7
8 1Corresponding Author Mobile, +91-9538020530; Email, sriranjini6@gmail.com
9
10 ABSTRACT
11 V.cholerae was first isolated by Italian anatomist Filipo Panici. V.cholerae is the etiological agent of cholera, a
12 major health concern in most of the developing countries. V cholerae carry strains the encode the cholera toxin.
13 These cholera toxins enters the Epithial cells and after crossing host line of defense it starts colonizing itself in
14 the small intestine. Cholera is usually a non contagious disease.
15 The main aim of this project is to design and develop a vaccine against cholera. Vibrio Cholerae is a bacterium
16 with 12,865 odd proteins causing cholera. Among these 1 protein sequence was selected having least identity
17 and least E- value. It was then screened by using SDSC workbench tool. Then antigenic determinants were
18 found by using different tools. The sequence with least identity was taken into consideration and then further
19 designed and used for docking studies. From this Docking analysis the epitope molecule LEALVEDL was
20 found to be the best vaccine candidate.
21
22 1. Introduction
23 Vibrio cholera also called as kommabacillus is a gram-negative comma shaped bacterium with a polar flagellum
24 that causes cholera in humans. V.cholera follows a fecal-oral infection path. It moves from contaminated food
25 and water and colonizes in the small intestine. V.cholera produces cholera toxin, the enterotoxin which acts on
26 the mucosal epithelium resulting in diarrhea.
27 There are two major biotypes of v.cholera i.e classical and ‘EI Tor’ which can be identified by
28 hemagglutination test. They occur both in marine and fresh water surfaces. It causes severe diarrheal disease in
29 humans by food and water. It is one of the most fatal illnesses known as diarrhea.
30 1.1 Clinical Symptoms:
31 ![]() Cholera symptoms include watery faces
Cholera symptoms include watery faces
32 ![]() With bits of mucus and mild fishy smell
With bits of mucus and mild fishy smell
33 ![]() Vomiting
Vomiting
34 ![]() Abdominal cramps
Abdominal cramps
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 7, June-2012 2
ISSN 2229-5518
35 ![]() Dehydration
Dehydration
36 ![]() Fever, it is rare, usually found in children.
Fever, it is rare, usually found in children.
37 The primary symptoms of cholera are profuse painless diarrhea and clear vomiting. These symptoms start
38 within 1 to 5 days after ingestion of bacteria. An untreated person may produce 10-15 lit. of diarrhea a day with
39 fatal results. If severe diarrhea and vomiting are not treated aggressively it may result in life threatening
40 dehydration and electrolytic imbalances.
41 1.2 Mode of transmission:
42
43 Transmission occurs through ingestion of contaminated water and food. Sudden large outbreaks are usually
44 caused by a contaminated water supply. Raw or undercooked seafood may be a source of infection in areas
45 where cholera is prevalent and sanitation is poor. Transmission due to direct person to person contact is rare.
46
47 2. Materials and Methods
48 2.1 Screening of Proteins
49
50 SDSC workbench: This bioinformatic tool is very essential in screening of proteins. By this screening the
51 protein with least identity was identified and its antigenic determinant was found.
52
53 The following protein ID with least identity was found are ;
54
55 Gene ID was found to be IF2_VIBC3 and its accession number is 172047700 with least identiy 20.62.
56
57 Least Identity – 21.89 , gene ID was found to be – 189046614 and the Accession number is
58 SECA_VIBC3
59
60 2.2 IMMUNOMED GROUP
61 This is mainly used to find out the antigenic determinants of the sequence that has least ident ity. From this tool
62 the Average antigenic propensity for protein sequence was found to be 1.0097
63
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 7, June-2012 3
ISSN 2229-5518
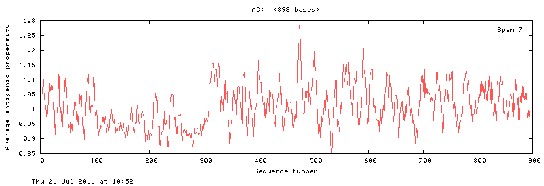
64
65 Fig 1. Average Antigenic Propensity
![]()
n | Start Position | Sequence | End Position |
1 | 4 | ITVKALS | 10 |
2 | 13 | IGTPVDRLLEQ | 23 |
3 | 42 | KQKLLAHL | 49 |
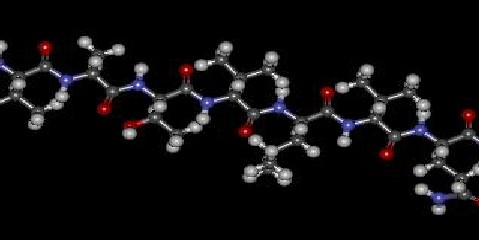
4 | 84 | KNVQVEV | 90 |
5 | 92 | KKRTYVKR | 99 |
6 | 208 | QLEKVRE | 214 |
7 | 237 | TDYHVTT | 243 |
8 | 308 | DKTAVVAKADVVVGETIVVSE | 328 |
9 | 336 | KATEVIK | 342 |
10 | 352 | TINQVID | 358 |
11 | 395 | EVSRAPVVTIMGHVDH | 410 |
12 | 420 | RRTHVAS | 426 |
13 | 432 | ITQHIGAYHV | 441 |
14 | 469 | ATDIVVLVVAA | 479 |
15 | 484 | MPQTVEAIQHAKAAGVPLIVAVN | 506 |
16 | 549 | IDGLLEAILLQAEVLELKAVKQ | 570 |
17 | 572 | MASGVVIE | 579 |
18 | 586 | RGPVATVLVQS | 596 |
19 | 601 | KGDIVLCGQEYGR | 613 |
20 | 629 | GPSIPVEILGLSGVPA | 644 |
21 | 647 | DEATVVR | 653 |
22 | 670 | REVKLAR | 676 |
23 | 692 | GDVALNIVLKADVQGSVEAIADSLTK | 717 |
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 7, June-2012 4
![]()
ISSN 2229-5518
24 | 719 | STDEVKVNIVGSGV | 732 |
25 | 737 | ETDAVLAAASNAIVVGF | 753 |
26 | 771 | DLRYYSIIYQLIDE | 784 |
27 | 798 | KQEIIGLAEVRDVFKSPKLGAIAGCMVTE | 826 |
28 | 833 | APIRVLRDNVVIY | 845 |
29 | 856 | KDDVAEV | 862 |
30 | 866 | YECGIGV | 872 |
31 | 876 | NDVRVGDQIEVFET | 889 |
66
67
68
69 2.3 MAPPP
70
Table 1
71 This tool is used to find out the type of MHC molecule (MHC 1 or 2) to which the epitope molecule binds.
72
73 Mappp results were found to be
74
Epitope | Position | MHC type | n- mer | Overall score | Cleavage Probability | MHC binding score | Group | |
0..897 | 898 | MTQITVKALSEEIGTPVDRL..VRVGDQIEVFETIEIQRTID | ||||||
QRKTRSTL | 66 | HLA_B8 | 8 | 0.8500 | 1.0000 | 0.7000 | n-term. trimmed | |
QRKTRSTL | 66 | HLA_B8 | 8 | 0.8500 | 1.0000 | 0.7000 | c-term. trimmed | |
QPRSDEEKL | 168 | HLA_B_0702 | 9 | 0.8429 | 1.0000 | 0.6857 | n-term. trimmed | |
QPRSDEEKL | 168 | H2_Ld | 9 | 0.8387 | 1.0000 | 0.6774 | n-term. trimmed | |
QPRSDEEKL | 168 | HLA_B_0702 | 9 | 0.8429 | 1.0000 | 0.6857 | n-term. trimmed | |
RRKAEEESR | 197 | HLA_B_2705 | 9 | 0.8514 | 1.0000 | 0.7027 | c-term. trimmed | |
PRGGKAGRK | 285 | HLA_B_2705 | 9 | 0.8514 | 1.0000 | 0.7027 | c-term. trimmed | |
KENELEEAI | 376 | H2_Kk | 9 | 0.8497 | 0.9994 | 0.7000 | trimmed twice | |
KENELEEAI | 376 | H2_Kk | 9 | 0.8498 | 0.9995 | 0.7000 | c-term. trimmed | |
ANPDNVKTEL | 512 | H2_Db | 10 | 0.9545 | 1.0000 | 0.9091 | n-term. |
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 7, June-2012 5
ISSN 2229-5518
trimmed | ||||||||
GLLEAILLQA | 550 | HLA_A_0201 | 10 | 0.8528 | 0.9997 | 0.7059 | same length | |
GLLEAILLQA | 550 | HLA_A_0201 | 10 | 0.8529 | 1.0000 | 0.7059 | c-term. trimmed | |
LLQAEVLEL | 556 | HLA_A_0201 | 9 | 0.9028 | 1.0000 | 0.8056 | n-term. trimmed | |
ILLQAEVLEL | 555 | HLA_A_0201 | 10 | 0.9559 | 1.0000 | 0.9118 | n-term. trimmed | |
EVLELKAVK | 560 | HLA_A3 | 9 | 0.8422 | 0.9868 | 0.6977 | c-term. trimmed | |
AIADSLTKL | 710 | HLA_A_0201 | 9 | 0.8916 | 0.9776 | 0.8056 | same length | |
AIADSLTKL | 710 | HLA_A_0201 | 9 | 0.9028 | 1.0000 | 0.8056 | c-term. trimmed | |
DEVKVNIV | 721 | H2_Kk | 8 | 0.8667 | 1.0000 | 0.7333 | c-term. trimmed | |
RYYSIIYQLI | 773 | H2_Kd | 10 | 0.9107 | 1.0000 | 0.8214 | c-term. trimmed | |
KRNAPIRVL | 830 | HLA_B_2705 | 9 | 0.8649 | 1.0000 | 0.7297 | n-term. trimmed |
75 Table 2
76 2.4 DISCOVERY STUDIO 2.5
77 MINIMIZATION OF MHC MOLECULES
78 The antigenic determinants LEALVEDL was designed
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 7, June-2012 6
ISSN 2229-5518
79 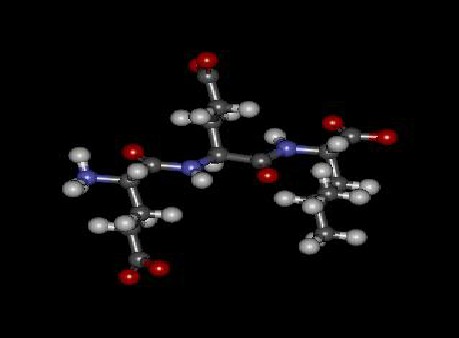
80 Fig 2. Epitope Molecule V.CHOLERAE
81 The minimization energy for this molecule is found to be 210.39238 kcal/mol.
82
83 The antigenic determinant GPVATVLVQSG was designed
84
85
86
87 2.5 Docking
88
89 This is mainly used to fit the epitope molecule into the MHC 1 molecule.
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 7, June-2012 7
ISSN 2229-5518
90 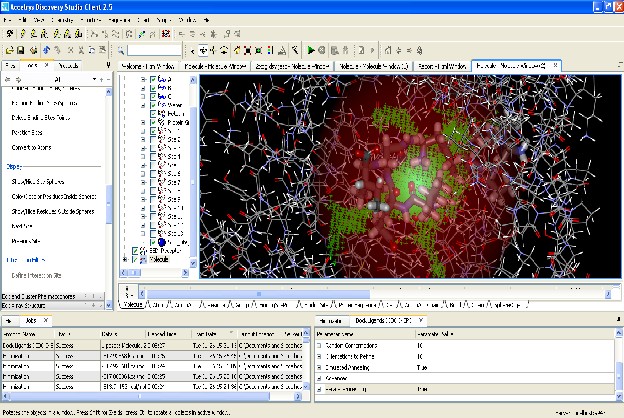
91 Fig 4. Docked Molecule V.CHOLERAE
92 INFERENCE : The epitope molecule was docked with MHC I molecule successfully
93 C docker Energy was found to be 74.1119
94 C docker Interaction Energy was found to be 41.0479
95
96 3. DISCUSSION
97
98
99
100
101
102
103
104
105
106
Thus from the above result it is found the antigenic determinant with least identity was ‘LEALVEDL’. The c
docker energy of this molecule was found to be 74.119 and C docker interaction energy was found to be
41.0479. By taking into consideration a vaccine is designed for cholera. The vaccine that is designed can be further sent for clinical trials and if it passes the FDA approval it can be used for curing this disease in future.
Support and help from the DNA Labs India, Hyderabad and Mr. Manas Ranjan Barik, is gratefully acknowledged.
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 7, June-2012 8
ISSN 2229-5518
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
1. Archivist (1997), Cholera phage discovery. Arch Dis Child 76 (3): 274. DOI:
10.1136/adc.76.3.274.Bentivoglio M, Pacini, P (1995). Filippo Pacini: A determined observer. Brain
Research Bulletin 38 (2): 161–5. DOI: 10.1016/0361-9230(95)00083-Q. PMID 7583342.
3. Bertranpetit J, Calafell F (1996). Genetic and geographical variability in cystic fibrosis: evolutionary
considerations. Ciba Found. Symposium. 197: 97–114; discussion 114–8. PMID 8827370.
4. DiRita VJ, Parsot C, Jander G, Mekalanos JJ (June 1991). Regulatory cascade controls virulence in Vibrio
cholerae. Proc. Natl. Acad. Sci. U.S.A. 88 (12): 5403–7. DOI: 10.1073/pnas.88.12.5403. PMC 51881. PMID 2052618.
5. Faruque, Shah M.; Nair, Balakrish, ed (2008). Vibrio cholerae: Genomics and Molecular Biology. Caister
Academic Press. ISBN 978-1-904455-33-2.
6. Faruque, SM; Nair, GB (2002). Molecular ecology of toxigenic Vibrio cholerae. Microbiology and immunology 46 (2): 59–66. PMID 11939579.
7. Fraser, Claire M.; Heidelberg, John F.; Eisen, Jonathan A.; Nelson, William C.; Clayton, Rebecca A.; Gwinn, Michelle L.; Dodson, Robert J.; Haft, Daniel H. et al. (2000). DNA sequence of both chromosomes
of the cholera pathogen Vibrio cholerae. Nature 406 (6795): 477–83. DOI: 10.1038/35020000. PMID 10952301.
8. Harris JB, Khan AI, LaRocque RC, et al. (November 2005). Blood group, immunity, and risk o f infection with Vibrio cholerae in an area of endemicity. Infect. Immun. 73 (11): 7422–7. DOI:
10.1128/IAI.73.11.7422-7427.2005. PMC 1273892. PMID 16239542.Hartwell LH, Hood L, Goldberg ML, Reynolds AE, Silver LM, and Veres RC (2004). Genetics: From genes to genomes. Boston: Mc-Graw Hill.
pp. 551–552, 572–574.
10. Howard-Jones, N (1984). Robert Koch and the cholera vibrio: a centenary. BMJ 288 (6414): 379–81.
DOI:10.1136/bmj.288.6414.379. PMC 1444283. PMID 6419937.
11. Karaolis, David K. R.; Somara, Sita; Maneval, David R.; Johnson, Judith A.; Kaper, James B. (1999). A
bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399 (6734): 375–9. DOI: 10.1038/20715. PMID 10360577.
12. King AA, Ionides EL, J.Luckhurst, Bouma MJ (August 2008). Inapparent infections and cholera dynamics.
Nature 454 (7206): 877–80. DOI:10.1038/nature07084. PMID 18704085.
13. Lan R, Reeves PR (Jan 2002). Pandemic Spread of Cholera: Genetic Diversity and Relationships within the Seventh Pandemic Clone of Vibrio cholerae Determined by Amplified Fragment Length Polymorphism (Free full text). Journal of Clinical Microbiology 40 (1): 172–181. DOI: 10.1128/JCM.40.1.172-181.2002. ISSN 0095-1137. PMC 120103. PMID 11773113.O'Neal C, Jobling M, Holmes R, Hol W (2005). Structural basis for the activation of cholera toxin by human ARF6-GTP. Science 309 (5737): 1093–6. DOI:
10.1126/science.1113398. PMID 16099990.
15. Ryan KJ, Ray CG (2004). Sherris Medical Microbiology (4th ed). McGraw Hill. pp. 376–7.
ISBN 0838585299.
16. Sack DA, Sack RB, Chaignat CL (August 2006). Getting serious about cholera. N. Engl. J. Med. 355 (7):
649–51. DOI: 10.1056/NEJMp068144. PMID 16914700.
17. Waldor, M. K.; Mekalanos, J. J. (1996). Lysogenic Conversion by a Filamentous Phage Encoding Cholera
Toxin. Science 272 (5270): 1910–4. DOI: 10.1126/science.272.5270.1910. PMID 8658163.
IJSER © 2012 http://www.ijser.org