International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013
ISSN 2229-5518
2343
CLASSIFICATION OF BRAIN ENCEPHALIC TISSUES FROM MRI IMAGES USING SPHERE SHAPED SUPPORT VECTOR MACHINE (SSSVM)
Ruta Sahasrabudhe, Prof. Megha Borse
Index Terms—3-D reconstruction, Region growing segmentation method, immune algorithm (IA), support vector machine
(SVM).
Human brain is the most vital and complex organ of our central nervous system. It is composed of 7 different types of tissues. It is really important to convert available 2-D MRI images to a 3-D brain to determine if there are any irregularities.3-D reconstruction of medical images is widely applied to tumor localization, surgical planning and removing the need for rescanning. The brain MRI images consist of wide range of the gray-scales and highly irregular boundaries. So it is difficult to classify different tissues using existing methods. These methods are tedious and time-consuming and most of the times don’t give satisfactory results.
Support vector machines (SVMs) are found to be very effective for the pattern recognition. In this paper, the sphere-shaped SVMs (SSSVMs) were used to for solving the classification problems due to class-imbalance. The main idea is that the SSSVM can transform an original 3-D object space into a high dimensional feature space by using kernel function. It constructs a compact hyper-sphere enclosing the entire target object in the feature space. The hyper-
sphere represents the target class and the points lying on or
Thus, the model of a 3-D object and data which support the near the surface of the hyper-sphere are Support Vectors. surface in the original space are obtained [4]. This method has several advantages. It can classify all the data points irrespective of how irregular their boundaries are and it also classifies several different types of data points of interest simultaneously and leads to near perfect reconstruction. This classification is achieved using kernel function. To make the model more flexible, parameters for SSSVM and kernel function are selected using Immune Algorithm (IA). So SSSVM along with IA (IASSSVM) is used for 3-D reconstruction.
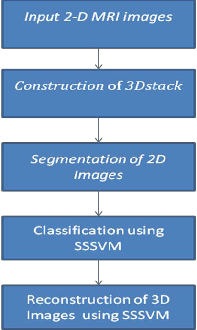
This paper is organized into seven sections. In section 1 block diagram of proposed system is explained. In section 2 image fusion technique is explained. Section 3 consists of explanation of region growing segmentation method. Section 4 explains Sphere Shaped SVM. In Section 5, Immune Algorithm is explained and Section VI comprises of conclusion and Results.
————————————————
Author is currently pursuing masters degree program in signal processing in Electronics & Telecommunications in Cummins College of Engineering, for Women, University of Pune, India, E-mail:ruta.halde@yahoo.com
Co-Author is currently Associate professor in Electronics & Telecommunications in Cummins College of Engineering, for Women, University of Pune, India, bmeghas@gmail.com
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013
ISSN 2229-5518
2344

Figure 1 Block Diagram of proposed system
Image fusion is the process of combining relevant information from two or more images into a single image. The resulting image will contain more information than any of the input images. For reliable and high quality image processing, high spatial and high spectral resolution should be present in single image. Most of the available equipment is not capable of providing such data. So image fusion is used which integrates different information sources.
It is very popular and used quite frequently in medical diagnostics and treatment. In this multiple patient images are registered and overlaid or merged to provide additional information. Images from same imaging modality (only MRI slices) can be fused together or fused images can also be created by combining information from multiple modalities, such as magnetic resonance image (MRI), computed tomography (CT), positron emission tomography (PET) etc. (WIKI)
The aim of data fusion is to use partial and varied information present in multiple images and creating a
single image with the collective features of all the input images. It exploits two main factors: one is the partial redundancy of data, since all sources provide information about the same phenomenon; the other is the complementarity between data since each source provides a different point of view about the phenomenon. Complementarity means certain features available in one object can be seen in a one image while it will be hidden in another one), as well as the type of information and the accuracy and quality of information (e.g. two different images of the same type but acquired with different parameters may be of different quality for different structures). Redundancy can be used in order to increase the global information and complementarity in order to improve certainty and precision.
Image segmentation is an important step for image processing. It is a process of partitioning a digital image into multiple regions (sets of pixels, super pixels). It simplifies and makes image more meaningful and easier to analyze. It is typically used to locate objects, boundaries (lines, curves, etc.) in images, to localize one or several different biological and anatomical regions of interest (ROIs) in the body [5].
Medical imaging is a most commonly used technique in
radiology to visualize the structure and function of the body. Successful numerical results in segmented medical image can help physicians and neurosurgeons to investigate and diagnose the structure and function of the body in both health and disease. Also, accurate segmentation is needed in clinical applications for doctors. But, segments in the medical images may not be manually obtained in accurate and efficient way. In other words, manual segmentation of medical images is a challenging and time-consuming task. Therefore, computer aided segmentation is very important to determine healthy results in medical images.
Region growing method is a simple and sophisticated method of image segmentation. It groups pixels or sub regions into larger regions based on predefined criteria
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013
ISSN 2229-5518
2345
[2].Uniformity criteria includes grey level uniformity, statistical and/or structure feature uniformity and texture feature uniformity [P3]. It involves the selection of initial seed points. The seeds mark each of the objects to be segmented. The regions are iteratively grown by comparing all unallocated neighboring pixels to the regions. The difference between a pixel's intensity value and the region's mean is used as a measure of similarity. The pixel with the smallest difference is allocated to the respective region. This process continues until all pixels are allocated to a region.
This method has several advantages - can correctly
separate the regions that have the same properties we define, possible to select our own seed points and criteria, possible to choose multiple criteria at the same time and performs well with respect to noise. Region growing methods can correctly separate the regions that have the same properties we define. Region growing methods can provide the original images which have clear edges with good segmentation results. The concept is simple. We only need a small number of seed points to represent the property we want, then grow the region. We can determine the seed points and the criteria we want to make .We can choose the multiple criteria at the same time. It also performs well with respect to noise.
SVMs (Support Vector Machines) are a useful technique for data classification. They are very specific class of algorithms, characterized by usage of kernels, absence of local minima, sparseness of the solution and capacity control obtained by acting on the margin, or on number of support vectors. They are based on the concept of decision planes that define decision boundaries. A decision plane is one that separates between a set of objects having different class memberships. As a classifier it separates a set of objects into their respective groups with a line. Most classification tasks, however, are not that simple, and often more complex structures are needed in order to make an optimal separation, i.e., correctly classify new objects (test cases) on the basis of the examples that are available (train cases).The basic idea is that it separates different classes in
such a way that the separation is almost linear. The original
objects are mapped, i.e., rearranged, using a set of mathematical functions, known as kernels.
Support vector machines (SVMs) are effective methods for the general purpose of pattern recognition. The sphere-shaped SVMs (SSSVMs) were proposed for solving the classification problems of the class-imbalance. The main idea is that the SSSVM can transform an original 3-D object space into a high dimensional feature space by the kernel function. It constructs a compact hyper-sphere which encloses the target object in the feature space. The hyper- sphere represents the target class and the points lying on or near the surface of the hyper-sphere are SVs. Thus, the model of a 3-D object and data which support the surface in the original space are obtained [4].Selecting appropriate parameters for the SSSVM and the kernel function can make the model more flexible and can help to obtain more accurate data description. SSSVM has several advantages:- First, through the flexible hyper-sphere in the feature space, a 3-D object model can be reconstructed no matter how arbitrarily irregular the object surface is in the original space. In addition, there is un-necessity for priori information regarding the shape or the topology of an object. Also classify data into more than one category
1) Assume a dataset of 3-D points representing the object.
2) A minimal volume hyper sphere that contains all or most of data points should be found.
3) The accuracy of SSSVM lies on the design of an appropriate kernel.
4) In most cases, the RBF kernel works very well on![]()
SSSVM which has form as following:
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013
ISSN 2229-5518
2346
5) Two parameters that need to be adjusted (σ And C)
to increase the accuracy of the SSSVM as classifier.
6) σ is a constant which controls the trade off between the volume of the hyper-sphere and the fraction of target points rejected by the hyper-sphere;
7) C controls the effective range of the kernel function.
8) Two kinds of the errors needed to be minimized - number of target points rejected and number of non-target points accepted (RSE).
9) Firstly the Modelling Accuracy (MA) is defined as
NA/L, where L-> Number of target points and NA-
> No. of target points accepted by the hyper- sphere.
10) RSE is defined as NNA/NL which is set to be 5%, where NNA->is the number of the non-target points and NL-> is the number of non-target points accepted by the hyper-sphere.
11) In order to maximize the MA, IA is used to search for the optimal σ and C.
12) These optimal σ and C are then used for
classification on different brain tissues.
5 IMMUNE ALGORITHM
The immune system is the basic and remarkable defensive system against bacteria, viruses and other disease-causing organisms. It can produce millions of antibodies from hundreds of antibody genes for protecting animals from infection by foreign substance. The immune system has led to artificial immune algorithm. This algorithm can be effectively used for pattern identification, learning, memorizing, adaptability, diversity, and distributed mechanisms as these are similar to immune phenomenon in nature.
Generally, the algorithm starts with the assumed
solution. This “solution” is not an “answer” to the problem
but is the course the system would follow to reach an
optimum solution. The optimum solution cannot be reached but will take several iterations to give near perfect solution.
STEPS:
The description of algorithm is clearly showed by the following steps:
1) Analyse problem and assume a suitable solution.
Antigen is the problem ready to be solved and the antibody is the solution to the problem.
2) Produce initial antibodies at random..
3) Compute the similarity among antibodies.
According to the similarity, suppress antibodies of high similarity as it leads to duplication.
4) Compute the affinity of antigen-antibody.
Compute the affinity of antigen-antibody.
5) Execute clone selection, that is remove antibodies of low affinity and clone antibodies of high affinity.
6) For this, each antibody set is input into the SSSVM
system respectively.
7) Their RSEs are computed firstly and the antibody sets are removed if the RSEs are higher than 5%.
8) Antigen is the problem ready to be solved and the
antibody is the solution to the problem.
9) The MAs can be obtained for the rest of antibody sets.
10) The antibody sets with lower than the mean MA
are removed
11) Antibody sets with higher than the mean MA are stored into the immune memory matrix.
12) This is initial antibody generation for the next iteration.
13) Produce next antibody generation by antibody mutation to ensure that optimizing process evolves to global optimum.
14) Through iteration, the best MA can be obtained if the MA can no longer be improved or the termination condition is reached.
15) The parameters corresponding to the best MA are
the optimal σ and C.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013
ISSN 2229-5518
2347
In this section the output using SSSVM as a classifier is presented. The Algorithm was implemented in Matlab®.R2011b.MRI images were acquired using a 3-D T2 weighted fast-spin echo pulse sequence with the voxel size of 1 X 1 X 1 mm with a matrix size of 256 X 256 X 150.The brain MR images include seven contrasted objects including the Background (BG) and six kinds of encephalic tissues, i.e., Scalp (SC), Osseous Compact Substance (OCS), Osseous Spongy Substance (OSS), Cerebral Spinal Fluid (CSF), Cerebral Gray Matter (CGM) and Cerebral White Matter (CWM). For training and testing purpose, in this study, the images are fused together figure 5 which shows fusion of slices 1,2,3 in figure 2,figure 3 and figure 4 and then these slices were segmented using Region growing method which gives better results in presence of noise and can segment more than one type of brain tissue thus saving lot of processing time and efforts. The output of this method is shown in figure 6 and figure 7.The classification is done using SSSVM along with IA to select appropriate parameters for Redial Basis Function(RBF).With this the possibility of wrong classification is considerably reduced which is the paramount consideration in any type of medical image processing. Figure 9 show all types of brain tissues classified using SSSVM in different color with great results.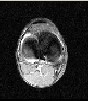
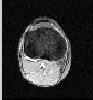
Figure 2: Slice 1 Figure 3: slice 2

Figure 4: Slice 3 Figure 5: Fusion of slices
1,2,3

Figure 6: Cerebral Figure 7: Cerebral grey
White Matter after matter after segmentation of
Segmentation of Slices 1,2,3
Slices 1,2,3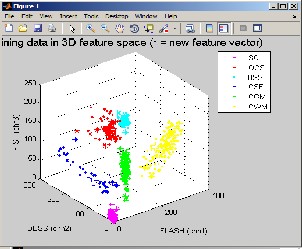
Figure 8 : Training data in 3D feature Space
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013
ISSN 2229-5518
2348
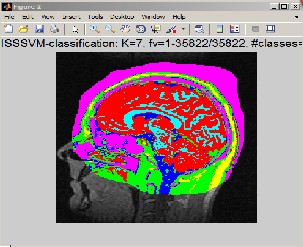
Figure 9: Mapping of classified tissues on brain map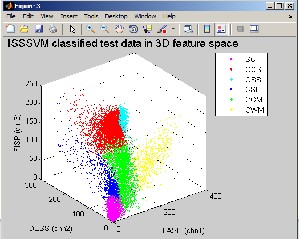
Figure 10: ISSSVM classified test data in 3D feature space
For each tissue, odd slices were used as the training set of the system and even slices were used to test the system performance. 1000 points from each object were chosen randomly as the training (validation) data set. The validation set was fed to the hyper-sphere of the target object to obtain its RSE. The proposed system was run on a personal computer programmed with MATLAB language. The computer CPU was 2.30 GHz with 4 GB of RAM. Also Adobe Flash Player should be installed so as to view the
classification progress clearly.The computational time
varies with the number of 3-D points of each object. The computational time depends on the size of training and testing data sets.
The accuracy of the developed system highly depends on the accuracy of the training data set processing and the similarity between the training data and testing data. Also it depends on the quality of MR images and good image contrasts among different objects in the MR images. In this case, it is found to be almost 98%.
In the brain MR images, due to highly irregular surface of each encephalic tissue it is difficult to do 3-D reconstruction of the brain using traditional techniques. So owing to the capability of SSSVM of solving non linear problems, it has been used. It also gives control over the parameters that defines the accuracy of the reconstruction. This parameter selection is done using Immune Algorithm which finds the optimal parameters. It is clear from the study in this paper that SSSVM is very successful in classifying the data with high irregularities. It is used to reconstruct objects with irregular boundaries with high precision. It has a great potential in medical imaging where almost all the data Is very irregular.
[1]Lei Guo, Ying Li, Dongbo Miao, Lei Zhao, Weili Yan, and XueqinShen , “3-D Reconstruction of Encephalic Tissue in MR Images Using Immune Sphere shaped SVMs”,IEEE Transactions On Magnetics, Vol. 47, No. 5, May 2011
[1] J. P. Chilès, C. Aug, A. Guillen, and T. Lees, in Proc.
Orebody Model.Strategic Mine Planning, Perth, Australia, Nov. 2004, pp. 313–320.
[2] G. Treece, R. Prager, and A. Gee, Vis. Comput., vol. 17, no. 7, pp.397–414, Oct. 2001.
[3] N. Cristianini and J. S. Taylor, An Introduction to Support
Vector Machines.Cambridge, U.K.: Cambridge Univ. Press,
2000.
[4] P. Y. Hao, J. H. Chiang, and Y. H. Lin, Appl. Intell., vol.
30, no. 2, pp.89–111, Apr. 2009.
[5] V. Cutello, G. Niscosia, M. Pavone, and J. Timmis, IEEE
Trans. Evol.Comput., vol. 11, no. 1, pp. 101–117, Feb. 2007.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013
ISSN 2229-5518
2349
[6] Q. Wu et al., IEEE Trans. Magn., vol. 41, no. 5, pp. 1912–
1915, May2005.
[7] Y. Zhou, “Research on recognition method based on artificial immunity,” Ph.D. dissertation, Beijing Univ. Sci. Technol., Beijing, China,2004.
[8] L. Guo, X. Liu, Y.Wu, W. Yan, and X. Shen, in Proc. IEEE
29th Eng.Med. Biol. Soc., Lyon,, France, Aug. 2007, pp. 6019–
6022.
[9] K Narayanan ,YogeshKarunakar, “3-D Reconstruction of Tumors in MRI Images”, International Journal of Research in Signal Acquisition and Processing,Vol.1,No.2,June
011,ISSN: 2046-617X
[10] N. MohdSaad , S.A.R. Abu-Bakar , SobriMuda , M. Mokji , A.R. Abdullah, “Automated Region Growing for Segmentation of Brain Lesion in Diffusion-weighted MRI”,Proceedings of the international MultiConference of Engineers and computer scientists 2012 Vol.1,IMECS
2012,March 14-16 2012, Hong Kong
.
IJSER © 2013 http://www.ijser.org