International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1788
PUDUCHERRY CAMPUS. PRIST UNIVERSITY PUDUCHERRY CAMPUS
Abstract: Collaboration has become a d ominant feature of modern science. Many scientific problems are beyond the realm of individual discipline or scientist to solve and hence require collaborative efforts. Meanwhile, today’s science becomes increasingly more data-intensive, resulting in a rapid transition from computational science to e-Science (or digital science). Recently, scientific workflows have emerged for scientists to integrate distributed computations, datasets, and analysis tools to enable and accelerate scientific discovery. The convergence of the above two trends naturally leads to the concept of collaborative scientific workflows. This paper presents a disciplinary definition of this term, discusses the opportunities, requirements, and challenges of collaborative scientific workflows for the enablement of scientific collaboration, and concludes with our ongoing work in this direction.
Keywords: Scientific collaboration, scientific workflows, collaborative scientific workflow
![]()
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1789
1. INTRODUCTION
![]()
In recent years, scientists have started to use scientific workflows to integrate and structure local and remote heterogeneous computational and data resources to perform in silico experiments and have made significant scientific discoveries. In contrast to business workflows that are controlflow oriented and orchestrate a collection of well- defined business tasks to achieve a business goal, scientific workflows are dataflow oriented and streamline a collection of scientific tasks to enable and accelerate scientific discovery [1, 2]. Several scientific workflow management systems (SWFMSs) have been developed to support scientific workflow design and execution, such as Kepler [1], Taverna [3], Triana [4], VisTrails [5], Pegasus [2], Swift [6], and VIEW [7, 8].
Existing SWFMSs mainly support single scientists to compose and execute scientific workflows. Modern scientific research projects, however, are collaborative in nature, and team members usually reside at geographically distributed locations. For example, the Cancer Biomedical Informatics Grid (caBIG) initiative launched by the
National Cancer Institute aims to connect the entire global cancer community to accelerate cancer research [9]. Therefore, there is a compelling need for a proper IT infrastructure and online services to support collaborative scientific workflows on the Internet. We define a collaborative scientific workflow as the computerized facilitation or automation of a scientific process, in whole or part, which streamlines and integrates people, datasets, and scientific tasks with data channels, dataflow constructs, and collaboration patterns to automate collaborative data computation and analysis for enabling and accelerating scientific discovery.
Building Internet-based services to support collaborative scientific workflows poses significant challenges. One main challenge is to understand the sophisticated interaction and hierarchical composition of various dataflow constructs and collaboration patterns to model complex and large-scale scientific workflows among scientists. A second challenge is to capture, manage, and utilize large amounts of distributed, heterogeneous, multi-level, and collaborative provenance data for the reproducibility of scientific results produced from collaborative scientific workflows.
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1790
scientists to collaboratively compose and manipulate a shared scientific workflow as the scenario shown in Fig. 1.
As a starting point, this paper examines the state of the art of the field of scientific workflows toward supporting collaborative scientific workflows that are targeted for collaborative science. Our preliminary research work is also reported to evaluate the trend toward the direction and inspire extensive research work. The remainder of the paper is organized as follows. Section 2 motivates our research. Section 3 di scusses existing work. Section 4 pr esents research challenges. Section 5 pr esents our preliminary work toward the direction of collaborative scientific workflows. Section 6 makes conclusions.![]()
2. MOTIVATION OF COLLABORATIVE SCIENTIFIC WORKFLOWS
![]()
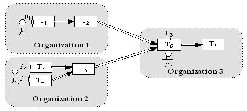
The latest advance of IT technologies have enabled and encouraged people to form large-scale and multidisciplinary scientific research projects to solve complex scientific problems. Demanding intensive computation and data sharing, these projects are collaborative in nature and usually include multiple domain scientists with domain- specific expertise located at geographically distributed organizations. Fig. 1 illustrates a scientific workflow comprising seven tasks (T1~T7) that have to be conducted by three scientists from three organizations at distributed locations. As shown in Fig. 1, t he tasks are not isolated from each other. Instead, they have to be streamlined in a defined workflow to produce useful scientific results. In other words, every scientific workflow run requires all three scientists to collaborate, either synchronously or asynchronously.
Fig. 1. A collaborative scientific workflow. Therefore, there is a compelling need for an online system
to support such collaborative scientific workflows on the Internet. Such a system should provide services to allow participating scientists to view the progress of the entire workflow, repeat a workflow run, and communicate and collaborate with peer scientists to perform scientific workflows. The system should also enable these projects to dynamically structure and integrate computations, datasets, scientists, and other resources or even workflows from multiple autonomous organizations with the goal of solving a scientific problem collaboratively.
Several scientific workflow management systems (SWFMSs) have been developed as single-user environments that focus on helping an individual scientist compose scientific workflows from available resources. Some systems show some collaboration features, in the sense that they allow a scientist to compose a scientific workflow from shared services, e.g., published Grid services. However, they provide limited support for multiple
They do not address and support user interaction and cooperation that are required and essential for an effective and efficient scientific collaboration.
In addition, current SWFMSs are built on top of different workflow models. Thus, their interoperability is poor. It is neither practical nor feasible to require that every domain scientist in a large-scale research project to adopt the same SWFMS and tool. Meanwhile, note that it is common for a domain scientist to participate in multiple scientific collaboration projects simultaneously. Therefore, it is critical to establish fundamental models to support collaborative scientific workflows, so that interoperability can be achieved among different SWFMSs.
Although the business world has recognized similar need [10] and has developed a preliminary model to support business workflows involving humans [10], the model is inapplicable to collaborative scientific workflows due to the fundamental differences between business workflows and scientific workflows: While business workflows are controlflow oriented, scientific workflows are dataflow oriented, introducing a new set of requirements for system development [11]. Moreover, provenance management has become a critical functionality for scientific workflows [12]. Although provenance bears much similarity to audit trails in business workflows, provenance provides much richer information than audit trails do. While audit trails only record temporal information concerning what operations are performed by whom at what times, provenance, in addition, records the causal relationships between these activities. Moreover, audit trails serve the purpose of auditing, while provenance is used for reproducibility of scientific results. In summary, the lack of collaboration support and interoperability among SWFMSs has largely limited the potential of using scientific workflows to enable and accelerate scientific discovery and to solve scientific problems that require collaborative efforts. If research on collaboration and interoperability lags implementation too much, IT scientists and engineers will have to retrofit techniques to achieve these requirements. They will have fewer options and most likely end up with a suboptimal solution.![]()
3. STATE OF THE ART OF SCIENTIFIC WORKFLOWS
![]()
To understand the challenges and opportunities of supporting collaborative scientific workflows, it is critical to examine the state of the art of the field of scientific workflows. Below, we focus on analyzing existing systems and their scientific workflow models, provenance models, and collaboration support.
Several scientific workflow management systems (SWFMSs) have been developed over the past decade. Their key features are summarized in Fig. 2. Kepler [1] is a Java- based open-source SWFMS, where a scientific workflow is composed of components called actors and its execution is controlled by a computational model controller called director. Taverna [3] is an open-source SWFMS targeted for life science. Based on a repository of services supporting
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1791
SWFMS | Key features |
Kepler | multiple models of computation; data in various formats; data, workflow, component |
Taverna | SCUFL for workflow representation; Web services support; GUI; social groups |
Triana | sophisticated graphical user interface |
VisTrails | visualization of evolving workflows; provenance management |
Pegasus | mapping workflows on Grid; manage workflow execution |
Swift | Grid-based workflows with short-running tasks; a parallel scripting language & execution engine |
VIEW | service-oriented architecture and efficient provenance querying and management |
Fig. 2. A comparison of existing SWFMSs.
various bioinformatics data analysis and transformation, Taverna uses an XML-based workflow language called SCUFL/XSCUFL for workflow representation with each component being either a Web service or a processor developed using Java Beanshell script. Taverna also features by its professional graphical user interface. Triana [4] provides a sophisticated graphical user interface for workflow composition and modification, including grouping, editing, and zooming functions. VisTrails [5] focuses on workflow visualizations supporting provenance tracking of workflow evolution in addition to data product derivation history. Pegasus [2] provides a framework that maps complex scientific workflows onto distributed Grid resources. Artificial intelligence planning techniques are used for guiding workflow composition. Using an actor- oriented modeling mechanism, Pegasus manages workflow execution and enables automatic retries at failures. Swift [6] combines a scripting language called SwiftScript with a runtime system to support specification and execution of large-scale loosely coupled computations over a Grid environment. Finally, our VIEW [7, 8] system features a service-oriented architecture and efficient provenance management for scientific data visualization [12].
Each of these SWFMSs provides a platform to support a single scientist to compose scientific workflows from various resources. Their foundations center on scientific workflow models and provenance models, which are reviewed below.
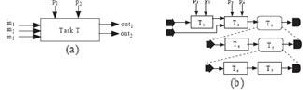
Tasks are considered basic building blocks of scientific workflows. Existing task models [1, 3] are illustrated in Fig. 3.(a), where a task represents a computational or analytical step of a scientific process. Each task comprises a set of input ports and output ports as its communication interface to other tasks. As shown in Fig. 3.(a), a task may also comprise an arbitrary number of input parameters (special kinds of input ports) that can be used by a scientist to configure the dynamic execution behaviors of the task.
Fig. 3. Scientific workflow model.
Centered on tasks, existing scientific workflow models [1, 3]
adopt a dataflow-driven modeling paradigm. As shown in Fig.
3.(b) as an example, tasks are linked together into a
workflow via data channels; a task will automatically start its execution whenever all required data become available at the input ports of the task. During workflow execution, tasks communicate with each other by passing data through data channels. As shown in Fig. 3.(b), a task in a workflow model may be a composite task that is expandable into a sub-workflow.
Some advanced techniques have been proposed to enhance the basic scientific workflow model. The transactional task model [13] uses concurrency control to ensure correct simultaneous access of databases and failure atomicity for workflow tasks. The shared hypermedia- based task model [14] supports simultaneous change, visualization, and navigation control of workflow task structure and attributes by multiple users.
In the current workflow models, however, human factor is not given sufficient consideration. The latest work of Taverna starts to investigate interaction patterns [15]. Simple parameter setting is supported at the task level. Recently, the WS-HumanTask model [16] is introduced to integrate humans into service-oriented business workflows. However, it d oes not support modeling of collaboration behaviors and patterns (e.g., user parameter control, steering control, and result validation control) that are required by a scientific workflow task. As shown in Fig.
3.(b), Tasks 1 t o 4 each belongs to its proprietary domain. The workflow model does not provide a facility to support the differentiation between the four domains and how they can collaborate on the workflow execution. Since many scientific process scenarios require constant, rich, and intricate user actions, it is important to develop a workflow model for collaborative scientific workflows that supports the flexible, efficient, and effective modeling and management of interaction, coordination, collaboration among workflows, tasks, datasets, organizations, groups, and individuals.
Provenance management has been acknowledged as a critical functionality for any SWFMS [12, 17-21]; see [22,
23] for surveys. Provenance supports the reproducibility of scientific results. Provenance data captures the derivation
history of a data product, including the original data sources, intermediate data products, and the steps that are applied to produce the data product. In other words, provenance captures the detailed scientific protocol that is needed to reproduce a scientific discovery. An execution of the workflow shown in Fig. 3.(b) produces a workflow run provenance shown in Fig. 4.(a), which is a graph consisting of two types of nodes: circles represent parameter values
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1792
R.DEEPAVARTHINI
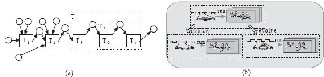
extract, and manage distributed provenance data to support the full lifecycle of a collaborative scientific workflow.
Fig. 4. Scientific workflow provenance.
and data products; rectangles represent task runs. Edges represent dependencies between nodes.
Kepler [1] implements a provenance recorder to track information about a workflow run, including its context, data derivation history, workflow definition, and workflow evolution. Taverna [3] uses Semantic Web technologies for representing provenance metadata at four levels: process, data, organization, and knowledge. VisTrails [24] records provenance for workflow evolution as well as for data product derivation and use it for collaborative design of scientific workflows [24]. Heinis and Alonso create an interval-based representation for provenance storage to save space [25]. Chapman et al. propose a set of factorization processes and inheritance-based methods to reduce the size of actual provenance datasets by up t o a factor of 20 [ 26]. To facilitate focused query and navigation over large amounts of provenance, Biton et al. develop a provenance abstraction technique called “user views” to return only relevant and abstracted provenance information to a user [27]. Our RDFProv system [12, 28] integrates the interoperability, extensibility, and reasoning advantages of Semantic Web technologies with the storage and querying power of a relational database management system and provides the first provably semantics-preserving SPARQL- to-SQL query mapping algorithms [29].
Several stand-alone provenance systems have also been developed [30-34], including the PReServ system developed under the Provenance Aware Service Oriented Architecture (PASOA) project [32] and the Karma system [33]. Both systems support Web services interfaces. To promote the interoperability of provenance among different systems, the Open Provenance Model was initiated in 2007 and has been positively influencing the community ever since [35]. In a collaborative scientific workflow environment, new requirements arise to provenance management due to the distributed nature of provenance data involving inter-domain data dependencies. As shown in Fig. 4.(b), the provenance data shown in Fig. 4.(a) is divided into two sub-graphs stored in two different provenance stores of the two domains representing participating organizations or security domains [36]. While some researchers have studied the problem of provenance capture for distributed scientific workflows [37], provenance management for collaborative scientific workflows, in which human collaboration and interaction are essential, has not been explored.
To support collaborative scientific workflows, a provenance model shall enable access and querying of provenance, across multiple workflow domains and at different levels. The goal is to support the reproducibility of scientific results obtained from the execution of collaborative scientific workflows performed by multiple scientists from different organizations. In addition, it is important to explore how to effectively and efficiently store,
Workflow scheduling is a major functionality of the workflow engine in an SWFMS [11], which schedules, assigns, and maps workflow tasks to machines or humans for execution. While early workflow scheduling work focuses on correctness issues, such as enforcing inter-task dependencies [38], recent work, mainly in the area of scientific workflow, focuses on performance, cost, and QoS based optimization of workflow scheduling [39].
A workflow scheduler can either be centralized, hierarchical, or decentralized. A centralized scheduler produces schedules for a whole workflow; a h ierarchical scheduler schedules a workflow by a cooperation of the root scheduler and its descendant low-level schedulers; a d ecentralized scheduler utilizes a network of sub-schedulers to make scheduling decisions of a workflow [39]. A scheduler can make a local scheduling decision, which is solely based on the information of the task and sub-workflow at hand, or a global scheduling decision, which is based on the information and structure of the entire workflow. A workflow schedule can be either static that is generated before workflow execution, or dynamic that is generated on-the-fly based on runtime dynamic information, or adaptive that is based on adaptation of a static workflow schedule toward runtime dynamic information. The goal of workflow scheduling is to optimize some objective function, such as overall workflow execution time (aka makespan), budget, deadline, or other QoS criteria. It has been shown that in general, the workflow scheduling problem in a distributed environment is NP-complete [40]. Thus, in practice, various heuristics-based algorithms are used.
Here, we briefly review a number of representative workflow scheduling algorithms (see [41] for a more comprehensive survey on this topic). The Min-Min algorithm determines for each task t of a given set of independent tasks, the resource r that provides the minimum estimated completion time (MECT), then schedules the task t with the minimum MECT on its corresponding resource r [42]. The Max-Min algorithm also calculates MECT for each task, but instead of choosing the task with the minimum MECT to schedule, it selects the one with the maximum MECT to maximize the overlap between long-duration task runs and short-duration task runs, thus reducing the overall execution time [42]. The sufferage algorithm [42] calculates MECT for each task t, as well as MECT2, which is the second best minimum ECT for the task, and their difference (MECT2-MECT) as the sufferage value, which quantifies the penalty of not assigning a task to its best resource. The task with the maximum sufferage value is chosen to schedule first onto the resource that provides the mimimum MECT. The DCA algorithm [43] provides a dynamic programming-based approach to optimize both overall execution time and economic cost. A user will designate one of them as the primary criterion and the other one as the secondary criterion. The user will also provide a sliding constraint for the primary criterion. The algorithm first optimizes solely for the first criterion, and
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1793
BI-CRITERIA SCHEDULING OF SCIENTIFIC GRID WORKFLOWS
then optimizes for the second criterion while preserving the primary criterion cost within the sliding constraint. The HEFT algorithm [44] analyzes the whole workflow and calculates a rank value for each task based on the critical path from the task to the exit task, which is based on the average execution time and average data transfer time of the tasks on all resources. All the tasks in the workflow is then ordered decreasingly and scheduled in that order by selecting the resources that can complete the tasks in the earliest time. The GRASP algorithm iteratively generates solutions and then keeps the best solution as the final schedule. Each iteration consists of a construction phase that generates a feasible solution, and a local phase that applies a local search to improve the solution. GRASP can generate better schedules than other scheduling techniques, as it searches the whole solution space with considering the whole workflow and all available resources, although scheduling overhead is subject to the cost of each iteration and the number of iterations.
None of the above workflow scheduling algorithms, however, considers human tasks and collaboration primitives that involve human collaboration and coordination, which are essential for collaborative scientific workflow scheduling. Therefore, workflow scheduling algorithms that minimize not only machine cycles, but also human cycles, as well as communication overhead introduced by data movement and human-human coordination should be investigated for collaborative scientific workflows.
Quality constraints are critical for workflow design and selection [45]. Cardoso et al. [46] elaborate theoretical concepts of a QoS-aware workflow on top of a set of metrics. Many researchers have been exploring how to model and manage QoS of workflows, not only to satisfy user requirements but also to enhance workflow adaptability to ever changing environments. QoS of workflows may be modeled in a comprehensive manner. Not only can it be modeled at different levels (e.g., task level or workflow level), but also may it comprise a variety of attributes (e.g., reliability, performance, and fault tolerance).
Based on the Multidimensional Multi-choice Knapsack Problem (MMKP), Kofler et al. [47] create a mathematical model to represent various QoS parameters of a workflow against configurable user requirements. They also develop a parallelizable branch and bound algorithm to maximize the measurement of workflow over the Kepler [1] system. Yu et al. [48] model QoS constraints in a combinatorial model (MMKP) and a graph model (Multi-Constraint Optimal Path (MCOP) problem).
Zeng et al. [49] propose a middleware to support QoS- aware workflow composition of Web services using the integer programming method. Their algorithms predict QoS values based on historical invocation records. Lv [50] proposes an evaluation algorithm to calculate three QoS values of a workflow based on the values of its comprising tasks: time, expense, and reliability.
Binder et al. [51] adopt Semantic Web technology to model user requirements, and propose a mathematical model that is equipped with genetic algorithms to calculate and optimize
workflow execution cost. Tao et al. [52] consider six QoS parameters: time, cost, reliability, availability, reputation, and security. They also propose a rotary hybrid discrete particle swarm optimization (RHDPSO) algorithm that disturbs double extremums to enhance premature convergence and local optimum.
Guo et al. [53] propose an XML-based language to define QoS requirements (i.e., performance) of a workflow and associate the document with a business workflow language BPEL [54] document to enable a QoS-aware workflow management system. Brandic et al. [55] extend BPEL with QoS extensions, and build a prototype of a QoS-aware workflow engine equipped with various planning strategies on the Grid. Patel et al. [56] adopt a document to specify QoS values to support workflow discovery and recommendation.
For business workflows, the term “collaborative workflows” is interchangeable with the term “coordinated workflows” [57-59]. They emphasize the coordination between workflows toward a common business goal.
The reference architecture proposed by the Workflow Management Coalition (WfMC) [60] has been widely adopted in the development of numerous business workflow management systems. Huang et al. [61] propose to use the agent technology to coordinate workflows. Dang et al. [62] propose to employ agents to coordinate workflow execution through a commitment-based formalisms by ontologically reasoning about their states and actions. Balasooriya et al. [63] propose a decentralized services-oriented middleware architecture to coordinate workflows. The central component of the architecture is a coordinator proxy object that manages dependencies of workflows and handles messages between the workflows. Balasooriya et al. [64] propose a two-layered framework, where distributed Web services are modeled as self-coordinating entities, and a workflow is created by interconnecting such entities into a distributed network of objects using Web bond primitives.
Some researchers particularly study workflow coordination in a Grid environment. Miller et al. [65] propose a protocol language to express and coordinate workflows comprising both reactive (Web services) and proactive (autonomous agents) tasks in a Grid environment. Prodan [66] studied how to efficiently execute the specification of workflow coordination on a Grid with
scalability. Based on γ-calculus, Nemeth et al. [67] model workflow coordination as molecules and reactions to enable autonomous evolution in a changing Grid environment.
The Computer Supported Cooperative Work (CSCW) community has also studied the workflow coordination problem. Compared to the distributed system-oriented workflow management that focuses on structured processes, CSCW-oriented workflow control focuses on the unstructured processing on the shared document by human collaborators. For example, collaborators with different ownerships possess different controls over the processes [68]. Therefore, a number of CSCW-oriented workflow systems automate the coordination and interoperation of workplace activities [69]. To name a few, Business Process Models (BPM) [70] is a collaborative business process modeling tool; OntoEdit [71] supports a concurrent
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1794
R.DEEPAVARTHINI
collaborative software engineering process; Yen et al. present a collaborative control design tool that allows privileged collaborators to change the process as needed [68]; OPCATeam [69] integrates the object-oriented and process-oriented paradigms into one single framework to enable the coexistence of structured processes and human interaction behaviors in one business process modeling system.
In contrast to collaborative business workflows that imply collaboration between business workflows, we use the term to refer to collaboration between workflow human users. The need for integrating human interaction and collaboration into a workflow model has been recently recognized in the business workflow community. Ayachitula et al. [72] divide workflows into human-centric workflows and automated process-based workflows. Russell et al. [73] propose to establish a separate team access control layer, which combines role and organization, to management access in a collaborative workflow environment. The BPEL4People [10] workflow model is proposed to extend the de facto industry standard business workflow language BPEL [54] to standardize the interaction between automated and human workflows. However, BPEL4People workflow model is not suitable to be used for supporting collaborative scientific workflows because:
1) BPEL is controlflow-oriented and hence lacks dataflow constructs for interaction, movement, and processing of large datasets; 2) every computational components in BPEL must be a Web service, thus, lacking the support of modeling user interaction and visualization intensive tasks.
Therefore, we argue that establishing a fundamental collaboration model is necessary. Such a collaboration model should be independent and can be plugged into other models dynamically to favor configurability and re- configurability. This requirement is critical for a collaborative SWFMS to become generally applicable to various scientific collaboration projects and domains. Different scientific research projects may adopt different collaboration policies. A basic collaboration model should be flexible enough so that it can be configured to support these diverse collaboration rules and patterns, and then be plugged into a generic scientific workflow management system to support the corresponding research projects.
A collaboration description language will be important to properly represent human interactions and collaboration rules and patterns. It can formalize the collaboration models for reasoning and tracking during collaborative scientific workflow design, execution, and management. It may further strengthen the collaboration model with higher flexibility and extensibility. Such a high-level collaboration description language can help participating scientists precisely define and regulate actions in the lifecycle of scientific workflows. Scientists involved in a collaborative scientific exploration are domain knowledge experts; however, they may not be software developers. Such a language can help those domain experts define a collaborative scientific workflow system, and an associated language compiler can automatically generate program code. Moreover, using a collaboration language to construct a collaborative scientific workflow application makes it possible for different scientific workflow systems to interact
and interoperate with each other to build new scientific workflows more easily and rapidly.
Existing scientific workflow languages and authoring tools are primarily designed to support automated scientific processes based on Web services and Grid services. Human user intervention and interactions are currently not supported. Due to the clear demand on supporting and standardizing human interaction and collaboration in scientific workflows, it is important to develop a collaboration language and authoring tool to support formal descriptions of scientific activities.![]()
4. REQUIREMENTS OF COLLABORATIVE SCIENTIFIC WORKFLOWS
![]()
An exploratory process, aiming for a scientific discovery and conducted by multiple distributed scientists through collaboratively running scientific workflows, can be modeled as an exploration of a provenance space as shown in Fig. 5. Conceptually, we model the provenance space in three dimensions: workflow evolution, parameter tuning, and input selection. Exploring along the workflow evolution dimension aims to design a new workflow that has the potential to lead to a scientific discovery; exploring along the parameter tuning dimension aims to determine the right parameter settings to enable such a scientific discovery; exploring along the input selection dimension aims to locate and select proper raw datasets in which the sought-after knowledge is hidden. In reality, parameter tuning and input selection may each become comprehensive and hierarchical for a particular workflow. For example, it is common that the parameter vector of a typical scientific workflow may contain dozens of various parameters.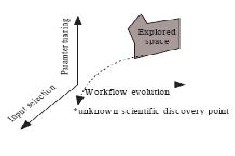
In such a provenance space, as shown in Fig. 5, the goal is
Fig. 5. The provenance space.
to run the exploratory process to find and reach a sought-after unknown scientific discovery point that corresponds to a proper scientific workflow, a suitable set of parameter settings, and an appropriate selection of input datasets. The strength of a collaborative scientific workflow is that the provenance space can be explored collaboratively and simultaneously by a group of geographically distributed scientists who share expertise, datasets, and other resources. For example, while one scientist is more experienced in tuning one set of parameters, another scientist may be more
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1795
BI-CRITERIA SCHEDULING OF SCIENTIFIC GRID WORKFLOWS
experienced in operating on a particular dataset.
The current scientific workflow infrastructure provides no or little support to expedite the collaborative exploratory process, thus resulting in a manual trial-and-error approach that has become one major bottleneck of today’s scientific discovery. Historic provenance data captures not only the execution and evolution history of workflows, but also the scientists’ computational thinking history represented as workflow designs, parameter tuning, workflow evolution, and input data selection. Our objective is to develop a procedure that automatically mines insightful knowledge from provenance data to assist scientists in future collaborative workflow evolution, parameter tuning, and input data selection.
Developing such a knowledge discovery process is highly difficult because: 1) The provenance space is infinite and the target scientific discovery points are unknown and may not be specified in advance or even foreseen; 2) Provenance data is high-dimensional, heterogeneous, and hierarchical, and hence provenance mining is not a trivial task; 3) The knowledge, constraints, and feedbacks from scientists have to be considered in a dynamic manner.

Unlike business collaboration focusing on the coordination of work (privileges and duties) among various parties [3], scientific collaboration concerns more about resource sharing at various levels [74]. We conducted a comprehensive study of the scientific collaboration literature, including a recent book entitled “Scientific Collaboration on the Internet” that is edited by Olson et al [75], which covers the challenges, experiences, and lessons summarized from a dozen of large-scale modern cyberinfrastructure projects, ranging from physical science, biological and health science, to earth and environmental science. Based on these studies and our research on scientific workflows and Internet-based collaboration, we identify eight key requirements for a collaborative SWFMS from a resource sharing perspective, as shown in Fig. 6.
Fig. 6. Requirements of scientific collaboration.
R1: Expertise sharing. The 21th century features the emergence of a new society, called the Knowledge Society, in which knowledge becomes the primary production resource instead of capital and labor. On one hand, the rapid growth of scientific knowledge increases the specialization of individual scientists. On the other hand, many of today’s complex scientific problems are beyond the realm of a
single discipline or scientist to solve. In the context of collaborative scientific workflows, a key requirement is easy and fast access to a pool of experts, so that various scientific tasks of a scientific workflow can be assigned to the scientists with proper expertise. Thus, a common collaboration pattern would be “to discover scientist S with expertise E to perform scientific task T of scientific workflow W.” Efficient and effective support of various expertise sharing patterns will not only form collaboration relationships that would be impossible otherwise, but also facilitate scientists in working together to answer questions that they cannot undertake alone.
R2: Workflow sharing. Modern scientific collaborations may require multiple scientists to collaboratively design, compose, execute, monitor, provenance track, and manage scientific workflows in both synchronous and asynchronous modes. Existing SWFMSs [3-7, 76] help individual scientists construct scientific workflows locally. Their individual work products (scientific workflows) are manually sent to collaborators (e.g., via emails) or uploaded into some shared social space (e.g., MyExperiment [77]) to enable collaboration. For example, collaborators can download a published Taverna workflow from MyExperiment, load it in to their local Taverna workbench, update, and send the new workflow back to the original collaborator for further changes. In the context of collaborative scientific workflows, an advanced collaborative environment is needed, where scientific workflows are encapsulated as services for publishing, discovery, and reuse.
R3: Data sharing. Modern science becomes more and more data-intensive, revealing the transition from computational science to e-Science (or digital science). Data sharing has become so important that some funding agencies, such as NIH, require that all large grant proposals must contain a data management plan to enable data sharing [78]. However, large volumes, complex types and structures, and the fear of losing data ownership pose great challenges to data sharing. In the context of collaborative scientific workflows, one key requirement would be the dynamic request of a desirable dataset from peers or collaborators. For example, during the execution of a scientific workflow, scientists may realize that a specific dataset is needed to continue the exploration, while the dataset belongs to an external group of scientists. Such a data sharing pattern should form a service interaction pattern so that both the data service requestor and the data service provider will achieve data sharing according to Service Level Agreements (SLAs). Moreover, data discovery will also be an important operation to support efficient and effective peer-to-peer (P2P) data sharing.
R4: Tool sharing. Although no tools can replace the critical and creative thinking and analysis of a scientist, software tools play an increasingly important role in scientists’ daily work in data processing and analysis. Sharing of these tools saves development expense and time; moreover, it encourages the reuse of a tool in a context that is beyond the imagination of the original developers. In the context of collaborative scientific workflows, supporting various tool sharing patterns is needed. For example, scientists may find that their obtained test data require
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1796
R.DEEPAVARTHINI
specific processing and analysis by a s pecific data analysis tool that is owned by, and maybe best operated by, an external group of scientists. One challenge for tool sharing is the shimming problem [79], which occurs due to the incompatibility of the interfaces and/or input/output data formats between external tools and existing tasks in a workflow.
R5: Computing power sharing. Nowadays, many scientific problems require the support of high-end computing, such as grid computing and cloud computing [80]. For example, in the ALICE project in Physics, it has been estimated that the project will require 35 M SI2k (1
MSI2k ≅ 430 CPUs) of computing capacity, 14 P B of disk
or transient storage, and 11 P B/year of tape or permanent
storage for best performance. In the context of collaborative scientific workflows, given the fast advance of high-end computing technology, a key requirement for computing power sharing is to separate the science-focused and technology-independent problem solving environment (PSE) from the underlying computing infrastructure. In this way, domain scientists can focus on their science while utilizing the state-of-the-art computing technologies in a transparent manner. Such a separation will also prevent vendor lock-in, so that users can seamlessly switch from one service provider to another.
R6: Storage sharing. The new generation of scientific experiments has resulted in a data deluge. For example, the Sloan Digital Sky Survey generates tens of terabytes of data. The proposal of the large Synoptic Survey Telescope is expected to produce more than 3 petabytes of data per year for the catalogs alone [75]. As a result, storage sharing becomes critical for processing and analyzing petascale scientific data. In the context of collaborative scientific workflows, storage needs to be abstracted as a utility based upon an on-demand basis. An architectural requirement is to enable a separation between a high-level data product model and a low-level storage model, so that the advancement of the storage model will not affect the data product model used in collaborative scientific workflow management.
R7: Instrument sharing. Some scientific problems solving requires the access of remote expensive physical instruments. The integration of the shared access of remote physical devices in a collaborative scientific workflow environment is a key requirement. For example, the Large Hadron Collider (LHC), the world’s largest and highest- energy particle accelerator at present, is a multibillion apparatus internationally shared in the high-energy physics community. Projects around LHC are highly collaborative: the ATLAS experiment alone involves over 1,800 physicists from 140 i nstitutions over 34 c ountries around the world [75]. How to manage and share such an expensive and sophisticated instrument is a big challenge, besides social and organizational challenges.
R8: Labor sharing. While scientific work features exploratory and creative, part of the work usually becomes routine and labor-intensive. For example, microarray data annotation and analysis are typically labor-intensive. Such work may be outsourced to a third-party organization, who can hire, train, and manage a group of data analysts more efficiently and effectively. The analysts can then become a pool for labor sharing. An efficient mechanism to facilitate
labor sharing in the context of collaborative scientific workflows may greatly reduce “human cycles” and accelerate scientific discovery, and might also reduce the cost of doing science.![]()
5. RESEARCH CHALLENGES
![]()
Based on our investigation of existing SWFMSs, we believe that it is important to develop fundamental models, language, architecture, and system to support collaborative scientific workflows. This section will discuss some key research challenges.
The major difficulties are three folds: 1) from a scientific workflow perspective, one has to understand and identify the requirements specific for scientific workflows, and then define and formalize various dataflow constructs in both syntax and semantics; 2) from a collaboration perspective, one has to understand various collaboration scenarios and requirements in the scientific workflow context, and then define and formalize various collaboration primitives and their composition properties; and 3) from an integrated model perspective, one has to investigate how collaboration primitives and dataflow constructs can be seamlessly integrated into one uniform collaborative scientific workflow model with well-defined and extensible semantics.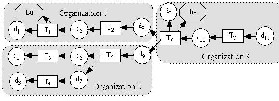
Fig. 7. Scientific workflow run provenance. Provenance of a scientific workflow captures the derivation
history of a data product, including the sources, intermediate data products, and the steps that are applied to produce the data product. Provenance management is essential for scientific workflows to support reproducibility
of scientific discovery, result interpretation, and problem diagnosis. Fig. 7 presents a scientific workflow run provenance for a particular execution of the scientific workflow shown in Fig. 1.
To ensure that scientific results are reproducible, a provenance system has to provide two key facilities. First, the provenance system has to collect and record sufficient provenance information, and support searching, querying, browsing, and visualization of provenance information. Second, the provenance system has to support the rerun or partial rerun of a workflow to reproduce or validate significant scientific results that are produced from running
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1797
BI-CRITERIA SCHEDULING OF SCIENTIFIC GRID WORKFLOWS
a collaborative scientific workflow.
Such a task is highly challenging due to the following characteristics of provenance data produced from the execution of collaborative scientific workflows: 1) Distributed: Collaborative scientific workflows typically involve resources from multiple organizations, as shown in Fig. 1. The capture and management of provenance are often distributed and inter-organizational provenance dependencies have to be properly modeled and managed. 2) Heterogeneous: Provenance produced from different scientific workflows often conforms to different schemas. Moreover, workflows may evolve rapidly, demanding a flexible provenance model to accommodate heterogeneity and evolution. 3) Hierarchical: Since scientific workflows are often constructed and managed hierarchically to deal with the complexity of scientific processes, the corresponding provenance data are multi-level. 4)
Such an effort should also explore and establish a methodology to guide domain scientists to construct project-specific collaborative scientific workflow systems. A reference architecture for building collaborative scientific workflow management systems based on aforementioned models should be provided, for the purpose of guiding a research project to easily customize and construct a domain-specific collaborative scientific workflow system, and enable interoperability between different collaborative scientific workflow systems. The sophisticated interaction and relationships between scientific workflow models and collaboration models require a deep investigation to design a sustainable architecture for various scientific collaborations.![]()
6. ONGOING RESEARCH
Collaborative: The collaborative provenance data model
has to be extensible to support new user interaction and collaboration patterns and store additional provenance information concerning the interactions and coordination
among scientists. Limited changes are allowed when additional user interaction and collaboration patterns are introduced.
5.3 Design of a Collaboration Model and a Collaboration
Scientific collaborations are typically featured as exploratory and unpredictable, and require constant user interaction and intervention in the process. For example, a scientific workflow may not be able to be fully composed at the beginning; participating scientists may discuss and creatively decide subsequent actions in the middle of the process based on intermediate experimental results; new collaborators may join in the middle of an exploration as the need for specific expertise and domain knowledge arises; participating scientists may possess different schedules and hence an asynchronous collaboration mode has to be supported in addition to a synchronous one; a scientific workflow may not have a clear time boundary and may last a long period of time, and so on. In addition, the introduction of human interaction may lead to other concerns such as co-design, co-run, co-monitor, and co- approve scientific workflows.
Thus, a collaboration model is required to capture and abstract such comprehensive and dynamic collaboration activities and patterns. Furthermore, such a collaboration model has to be flexible to endure constant changes and reconfigurations. For example, it is common that a scientist simultaneously participates in several collaborative scientific explorations, each specifying project-wise collaboration rules and protocols. Moreover, how to seamlessly incorporate a collaboration model with a task model, a workflow model, and w provenance model remains another challenge.
Toward the ultimate goal of developing a fundamental and generally applicable infrastructure to support the design, execution, monitoring, provenance tracking, and management of collaborative scientific workflows, we have conducted some preliminary research work. In this section, we report our research work in four directions: the VIEW and Confucius systems, the RDFProv provenance system, the CODL/XCODL collaboration languages, and an SOA- based infrastructure. Our ongoing work focuses on addressing the R2 (Workflow sharing) and R3 (Data sharing).
We have developed VIEW [7, 8], a service-oriented scientific workflow management prototype system. VIEW comprises a workbench to visually design workflows, a workflow engine to execute workflows [81], a provenance manager to store and query workflow provenance [12], and a data product manager to store and manage data products [82]. Using VIEW, a s cientist can create a new project consisting of multiple related scientific workflows. The workbench collects all workflow specifications into a log and, once the workflow design is complete, stores the workflow specifications through the record interface of the provenance manager to support the storage of workflow evolution provenance [5]. An existing workflow can evolve into a new workflow, augmenting its corresponding provenance database with new data. While a workflow runs, the workflow engine collects its execution provenance into a log and, once the execution finishes, stores it into the provenance database.
We have also developed Confucius 1.0 [83, 84], a prototyping system supporting collaborative composition of scientific workflows, built upon the Taverna [3] system. Using a client/server model, multiple scientists may join in a shared session to design scientific workflows collaboratively. Any change (adding or removal of components) made by one scientist will be immediately reflected on all collaborators’ screens.
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1798
R.DEEPAVARTHINI
We have developed a provenance prototype system, called RDFProv [28, 85], for storing and querying scientific workflow provenance data. Our approach combines the advantages of interoperability, extensibility, and reasoning of Semantic Web technologies with the power of storage and querying of a relational database management system. While the Resource Description Framework (RDF) model represents data in graphs, the relational model represents data in tables. To transform data in the RDF model to data in the relational model, the following three mappings were performed: 1) Ontology to relational database schema mapping, which takes an input scientific workflow provenance ontology and automatically generates a relational database schema that is optimized for common provenance queries; 2) Provenance data to relational data mapping, which maps a provenance dataset in RDF format to relational tuples conforming to the database schema generated by the first algorithm; and 3) SPARQL-to-SQL query mapping, which maps an input SPARQL query into an equivalent SQL query.
We also developed a relational operator, called nested optional join (NOJ) [85], to optimize SPARQL queries to enhance provenance query performance. By benchmarking the performance of RDFProv and other optimization strategies designed for scientific workflow provenance [28,
85], we proved that our solution provides higher efficiency and scalability to provenance data management [28].
We have developed a description language, called COllaboration Description Language (CODL) [86], to help the specification of the requirements of a collaboration in
the format of an electronic conference. A library of
collaboration primitives was constructed based on our study of proven human collaboration rules (i.e., Robert’s Rules of Order) and our extended procedure rule set adapted for
network and parallel operations [87]. A CODL runtime was developed to automatically translate CODL specifications into a set of collaboration primitives executed over a Java Virtual Machine. Based on CODL, we established a Member-Session-Meeting-Group architectural model to enhance collaboration control management [87]. A rule- mitigated synchronous collaboration environment was developed to permit users to gather in virtual meetings for discussion and decision making [87].
To eliminate the procedural style of CODL and permit flexible and dynamic specifications of coordination requirements, we extended CODL into X-CODL [88]. X- CODL serves as a plug-in to CODL as a description language focusing on defining coordination requirements. X-CODL models collaboration-oriented coordination requirements, while decoupling the coordination statements from collaboration business logic. We also developed a methodology to translate X-CODL specifications into Colored Petri Nets for simulation, analysis, and validation.
Based on our study and exploration, we found that Service
Oriented Architecture (SOA) [89] can play an important
role for constructing a generally applicable collaborative scientific workflow management system. Derived from the SOA Reference Architecture (SOA-RA) [90, 91] that provides a high-level template for developing SOA-based solutions with an abstraction of an SOA factored into layers, we define a two-dimensional layered architecture supporting scientific collaborations.
As shown in Fig. 8, the horizontal layers support application-specific functional requirements, and the vertical layers provide system-support facilities and enablement. Collaboration is separated from scientific workflow management into two layers. Provenance data models and management models are handled by a dedicated Data Architecture layer. Each layer shall comprise a set of design decisions, options, and key performance indicators. Provenance tracking of the execution of collaborative scientific workflows is performed in the Governance layer. The rerun or partial rerun of composed scientific workflows will be managed by the Scientific Workflow Management layer. Our SOA-based framework supports collaborative workflows. As shown in Fig. 8, vertical layers provide system-level support for distributed domain scientists to collaborate effectively and efficiently.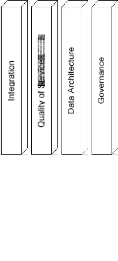

Fig. 8 SOA-based system architecture.
Our previous work introduces the concept of architectural building blocks (ABBs) that represent the constituent elements of a layer of SOA-RA [89]. For the SOA-based infrastructure to guide construction of collaborative scientific workflow management systems, we plan to examine our ABB pool and define ABBs for each layer in our architectural model.![]()
7. CONCLUSIONS
![]()
With the advent of the national cyberinfrastructure act and its focus on providing unprecedented IT support for scientific activities, research on mechanisms for automating and accelerating collaborative scientific discovery processes for a wide range of science and engineering disciplines has become more important than ever. In this paper, we survey the state of the art of the field of scientific workflows and identify the importance and research challenges of collaborative scientific workflows. We conclude that it is critical to develop a fundamental and generally applicable infrastructure to support the lifecycle of collaborative scientific workflows. We expect that SOA will play an
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1799
BI-CRITERIA SCHEDULING OF SCIENTIFIC GRID WORKFLOWS
essential role in designing and developing a generally applicable collaborative scientific workflow management system that can support resource sharing at various levels in the form of services.
17. Zhao, J., et al., Mining Taverna's Semantic Web of Provenance. Concurrency and Computation: Practice and Experience (CONCURRENCY), 2008. 20(5): p. 463-472.
18. Anand, K., S. Bowers, and B. Ludäscher. Techniques for Efficiently Querying Scientific Workflow Provenance Graphs. in EDBT. 2010.
19. Ellqvist, T., et al. Using Mediation to Achieve
ACKNOWLEDGEMENT
![]()
This work is supported by National Science Foundation, under grants NSF IIS-0959215 and IIS-0960014.
Provenance Interoperability. in SERVICES-I. 2009.
20. Kim, J., et al., Provenance Trails in the Wings-Pegasus System. Concurrency and Computation: Practice and Experience, 2008. 20(5): p. 587-597.
21. Clifford, B., et al., Tracking Provenance in a Virtual Data
Grid. Concurrency and Computation: Practice and
Experience, 2008. 20(5): p. 565-575.
REFERENCES
![]()
1. Ludäscher, B., et al., Scientific workflow management and
the Kepler system. Concurrency and Computation: Practice and
Experience, 2006. 18(10): p. 1039-1065.
2. Gil, Y., et al., Artificial Intelligence and Grids: Workflow Planning and Beyond. IEEE Intelligent Systems, 2004. 19(1): p. 26–33.
3. Oinn, T., et al., Taverna: Lessons in Creating a Workflow Environment for the Life Sciences. Concurrency and Computation: Practice & Experience, 2006. 18(10): p. 1067–1100.
4. Churches, D., et al., Programming Scientific and Distributed Workflow with Triana Services. Concurrency and Computation: Practice & Experience, 2006. 18(10): p. 1021–1037.
5. Freire, J., et al., Managing Rapidly-Evolving Scientific
Workflows Lecture Notes in Computer Science, 2006.
4145/2006: p. 10–18.
6. Zhao, Y., et al. Swift: Fast, Reliable, Loosely Coupled Parallel Computation. in IEEE International Workshop on Scientific Workflows. 2007. Salt Lake City, UT, USA.
7. Chebotko, A., et al. VIEW: A Visual Scientific Workflow Management System. in the 1st IEEE International Workshop on Scientific Workflows. 2007. Salt Lake City, UT, USA.
8. Lin, C., et al. Service-Oriented Architecture for VIEW: A Visual Scientific Workflow Management System. in the IEEE
2008 International Conference on Services Computing (SCC).
2008. Honolulu, HI, USA.
9. NCI. Cancer Biomedical Informatics Grid (caBIG). Available from: https://cabig.nci.nih.gov/.
10. Agrawal, A., et al. WS-BPEL Extension for People
(BPEL4People), Version 1.0. 2007 Jun.; Available
from: http://download.boulder.ibm.com/ibmdl/pub/software/dw/specs/
ws -bpel4people/BPEL4People_v1.pdf.
11. Lin, C., et al., A Reference Architecture for Scientific Workflow Management Systems and the VIEW SOA Solution. IEEE Transactions on Services Computing (TSC), 2009. 2(1): p. 79-92.
12. Chebotko, A., et al., RDFProv: A Relational RDF Store for Querying and Managing Scientific Workflow Provenance. Data and Knowledge Engineering (DKE), 2010. 69(8): p. 836-865.
13. Worah, D. and A. Sheth, Transactions in Transactional Workflows. Advanced Transaction Models and Architectures, 1997: p. 3-34.
14. Rubart, J. and H. Richter, Flexible Notifications and Task Models for Cooperative Work Management. Metainformatics, 2004: p. 32–41.
15. Lanzen, A. and T. Oinn, The Taverna Interaction Service: Enabling Manual Interaction in Workflows. Bioinformatics Applications Note, 2008. 24(8): p. 1118-1120.
16. Agrawal, A., et al. Web Services Human Task (WS-HumanTask), Version 1.0. 2007 Jun.; Available
from: http://download.boulder.ibm.com/ibmdl/pub/software/dw/specs/
ws -bpel4people/WS-HumanTask v1.pdf.
22. Bose, R. and J. Frew, Lineage Retrieval for Scientific Data
Processing: A Survey. ACM Comput. Surv., 2005. 37(1): p. 1-28.
23. Simmhan, Y., B. Plale, and D. Gannon, A Survey of Data
Provenance in e-Science. SIGMOD Record, 2005. 34(3): p. 31–36.
24. Freire, J., et al., Provenance for Computational Tasks: A Survey. Computing in Science and Engineering (CSE),
2008. 10(3): p. 11-21.
25. Heinis, T. and G. Alonso. Efficient Lineage Tracking for Scientific Workflows. in ACM International Conference on Management of Data (SIGMOD). 2008. Vancouver, Canada.
26. Chapman, A., H.V. Jagadish, and P. Ramanan. Efficient Provenance Storage. in ACM International Conference on Management of Data (SIGMOD). 2008. Vancouver, Canada.
27. Biton, O., et al. Querying and Managing Provenance through User Views in Scientific Workflows. in IEEE 24th International Conference on Data Engineering (ICDE). 2008. Cancun, Mexico.
28. Chebotko, A., et al. Storing and Querying Scientific Workflow Provenance Metadata Using an RDBMS. in the 3rd IEEE International Conference on e-Science and Grid Computing.
2007. Bangalore, India.
29. Chebotko, A., S. Lu, and F. Fotouhi, Semantics Preserving SPARQL-to-SQL Query Translation. Data & Knowledge Engineering, 2009. 68(10): p. 973-1000.
30. Cohen, S., S. Boulakia, and S. Davidson, Towards a Model of Provenance and User Views in Scientific Workflows. Lecture Notes in Computer Science: Data Integration in the Life Sciences, 2006. 4075: p. 264–279.
31. Bowers, S., et al. A Model for User-Oriented Data Provenance in Pipelined Scientific Workflow. in the International Provenance and Annotation Workshop (IPAW). 2006. Chicago, IL, USA.
32. Groth, P., et al. Recording and Using Provenance in a Protein Compressibility Experiment. in the 14th IEEE International Symposium on High Performance Distributed Computing
(HPDC). 2005. Washington, DC, USA.
33. Simmhan, Y., B. Plale, and D. Gannon. A Framework for Collecting Provenance in Data-Centric Scientific Workflows. in IEEE International Conference on Web Services (ICWS).
2006. Chicago, IL, USA.
34. Ellkvist, T., et al., Using Provenance to Support Real- Time Collaborative Design of Workflows Lecture Notes in Computer Science, 2008. 5272: p. 266-279.
35. The Open Provenance Model. Available from: http://openprovenance.org/.
36. Kang, M., J. Park, and J. Froscher. Access Control Mechanisms for Inter-Organizational Workflow. in the 6th ACM Symposium on Access Control Models and Technologies.
2001. Chantilly, VA, USA.
37. Marinho, A., et al. A Strategy for Provenance Gathering in Distributed Scientific Workflows. in SERVICES-I. 2009.
38. Attie, P.C., et al., Scheduling Workflows by Enforcing Intertask Dependencies. Distributed Systems Engineering (DSE), 1996. 3(4): p. 222-238.
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1800
Government (EEE). 2005. Las Vegas, USA.
39. Yu, J. and R. Buyya, A Taxonomy of Scientific Workflow
Systems for Grid Computing. SIGMOD Record (SIGMOD),
2005. 34(3): p. 44-49.
40. Fernández-Baca, D., Allocating Modules to Processors in
a Distributed System. IEEE Trans. Software Eng. (TSE), 1989.
15(11): p. 1427-1436.
41. Yu, J., R. Buyya, and K. Ramamohanarao, Workflow Scheduling Algorithms for Grid Computing. 2008, in Book “Studies in Computational Intelligence”, Springer Berlin/Heidelberg. p. 173-214.
42. Maheswaran, M., et al. Dynamic Matching and Scheduling of a Class of Independent Tasks onto Heterogeneous Computing Systems. in Heterogeneous Computing Workshop. 1999.
43. Wieczorek, M., et al. Bi-criteria Scheduling of
Scientific Workflows for the Grid. in CCGRID. 2008.
44. Topcuoglu, H., S. Hariri, and M.-Y. Wu, Performance- Effective and Low-Complexity Task Scheduling for Heterogeneous Computing. IEEE Trans. Parallel Distrib. Syst. (TPDS), 2002. 13(3): p. 260-274.
45. Chen, J. and Y. Yang, Adaptive Selection of Necessary and Sufficient Checkpoints for Dynamic Verification of Temporal Constraints in Grid Workflow Systems. ACM Transactions on Autonomous and Adaptive Systems, 2007. 2(2).
46. Cardoso, J., A. Sheth, and J. Miller, Workflow Quality of
Service, Enterprise Inter- and Intra-Organisational Integration
- Building International Consensus. 2002: Kluwer Academic
Publishers.
47. Kofler, K., I.u. Haq, and E. Schikuta. A Parallel Branch and Bound Algorithm for Workflow QoS Optimization. in 2009
International Conference on Parallel Processing. 2009.
48. Yu, T., Y. Zhang, and K.-J. Lin, Efficient Algorithms for
Web Services Selection with End-to-End QoS Constraints. ACM Transactions on the Web, 2007. 1.
49. Zeng, L., et al., QoS-Aware Middleware for Web Services
Composition. IEEE Transactions on Software Engineering,
2004. 30(5): p. 11–327.
50. Lv, T. Research on Workflow QoS. in 2009 International Joint
Conference on Artificial Intelligence. 2009.
51. Binder, W., et al. Optimal Workflow Execution in
Grid Environments. in NODe/GSEM.
52. Tao, Q., et al. QoS Constrained Grid Workflow Scheduling Optimization Based on a Novel PSO Algorithm. in 2009 8th International Conference on Grid and Cooperative
Computing. 2009.
53. Guo, L., et al. Enabling QoS for Service-Oriented Workflow
on GRID. in 7th IEEE International Conference on Computer and
Information Technology (CIT). 2007.
54. Jordan, D. and J. Evdemon. Web Services Business Process
Execution Language, Version 2.0. 2007 Apr.; Available
from: http://docs.oasis-open.org/wsbpel/2.0/OS/wsbpel-v2.0- OS.html.
55. Brandic, I., et al. QoS Support for Time-Critical Grid Workflow Applications. in 1st International Conference on e-Science and Grid Computing (e-Science). 2005.
56. Patel, Y., A. McGough, and J. Darlington. QoS Support For Workflows In A Volatile Grid. in 7th IEEE/ACM International Conference on Grid Computing. 2006.
57. Fakas, G. and B. Karakostas, A Workflow Management System Based on Intelligent Collaborative Objects. Information & Software Technology, 1999. 41(13): p. 907-915.
58. Song, H., et al. A SLA-Adaptive Workflow Integrated Grid Resource Management System for Collaborative Healthcare Services. in the 3rd International Conference on Internet and Web Applications and Services (ICIW). 2008. Athens, Greece.
59. Pudhota, L. and E. Chang. Collaborative Workflow Management Using Service Oriented Approach. in International Conference on E-Business, Enterprise Information Systems, E-
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1801
R.DEEPAVARTHINI
60. Hollingsworth, D., The Workflow Reference Model. The
Workflow Management Coalition, 1994.
61. Huang, C.-J., C.V. Trappey, and C.C. Ku. A JADE-Based Autonomous Workflow Management System for Collaborative IC Design. in the 11th International Conference on Computer Supported Cooperative Work in Design (CSCWD). 2007. Melbourne, Australia.
62. Dang, J., J. Huang, and M.N. Huhns. Workflow Coordination for Service-Oriented Multiagent Systems. in the 6th International Joint Conference on Autonomous Agents and Multiagent Systems (AAMAS). 2007.
63. Balasooriya, J., S.K. Prasad, and S.B. Navathe. A Middleware Architecture for Enhancing Web Services Infrastructure for Distributed Coordination of Workflows. in IEEE International Conference on Services Computing (SCC). 2008.
64. Balasooriya, J., et al. A Two-Layered Software Architecture for Distributed Workflow Coordination over Web Services. in IEEE International Conference on Web Services (ICWS). 2006.
65. Miller, T., et al. First-Class Protocols for Agent-Based Coordination of Scientific Instruments. in 16th IEEE International Workshops on Enabling Technologies: Infrastructure for Collaborative Enterprises (WETICE). 2007.
66. Prodan, R. Specification-Correct and Scalable Coordination of Scientific Applications in Grid Environments. in Seventh IEEE International Symposium on Cluster Computing and the Grid (CCGrid). 2007.
67. Nemeth, Z., C. Perez, and T. Priol. Distributed Workflow Coordination: Molecules and Reactions. in 20th IEEE International Parallel & Distributed Processing Symposium. 2006.
68. Yen, C., W.J. Li, and J.C. Lin, A web-based collaborative, computer-aided sequential control design tool. IEEE Control Systems Magazine, 2003. 23(2): p. 14-19.
69. Dori, D., D. Beimel, and E. Toch. OPCATeam - Collaborative Business Process Modeling with OPM. in 2nd International Conference on Business Process Management (BPM). 2004. Potsdam, Germany: Springer-Verlag Berlin Heidelberg.
70. Kazanis, P. and A. Ginige. Asynchronous collaborative business process modeling through a web forum. in Seventh Annual CollECTeR Conference on Electronic Commerce. 2002. Melbourne, VIC, Australia.
71. Sure, Y., et al. OntoEdit: collaborative ontology engineering for the semantic Web. in the First International Semantic Web Conference
2002 (ISWC), LNCS 2342. 2002: Springer.
72. Ayachitula, N., et al. IT Service Management Automation - A Hybrid Methodology to Integrate and Orchestrate Collaborative Human Centric and Automation Centric Workflows. in IEEE International Conference on Services Computing (SCC). 2007. Salt Lake City, UT, USA.
73. Russell, D., P.M. Dew, and K. Djemame. Service-Based Collaborative Workflow for DAME. in IEEE International Conference on Services Computing (SCC). 2005. Orlando, FL, USA.
74. Olson, G.M., A. Zimmerman, and N. Bos, eds., Scientific Collaboration on the Internet. 2008: MIT Press, Cambridge, MA, USA.
75. Olson, G.M., A. Zimmerman, and N. Bos, Scientifc
Collaboration on the Internet. 2008: The MIT Press.
76. Ludäscher, B., et al., Scientific Workflow Management and the Kepler System. Concurrency and Computation: Practice & Experience, 2006. 18(10): p. 1039-1065.
77. Roure, D.D., C. Goble, and R. Stevens, The Design and Realisation of the myExperiment Virtual Research Environment for Social Sharing of Workflows. Future Generation Computer Systems,
2009. 25: p. 561-567.
78. Lynch, C. and J. Kippincott, Insitutional Repository Deployment in the United States as of Early 2005. D-Lib Magazine, 2005. 11(9).
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 2, February-2014
ISSN 2229-5518 1802
BI-CRITERIA SCHEDULING OF SCIENTIFIC GRID WORKFLOWS
79. Lin, C., et al. A Task Abstraction and Mapping Approach to the Shimming Problem in Scientific Workflows. in IEEE International Conference on Services Computing (SCC). 2009. Bangalore, India.
80. Foster, I., et al. Computing and Grid Computing 360-Degree
Compared. in IEEE Grid Computing Environments. 2008.
81. Fei, X., S. Lu, and C. Lin. A MapReduce-Enabled Scientific Workflow Composition Framework. in IEEE International Conference on Web Services (ICWS). 2009. Los Angeles, CA, USA.
82. Fei, X. and S. Lu. A Collectional Data Model for Scientific Workflow Composition. in the IEEE International Conference on Web Services (ICWS). 2010. Miami, FL, USA.
83. Zhang, J., D. Kuc, and S. Lu. Confucius: A Scientific
Collaboration System Using Collaborative Scientific Workflows.
in IEEE International Conference on Web Services (ICWS). 2010. Miami, FL, USA.
84. Zhang, J. Co-Taverna: A Tool Supporting Collaborative Scientific Workflows. in IEEE International Conference on Services Computing (SCC). 2010. Miami, FL, USA.
85. Chebotko, A., et al., Relational Nested Optional Join for Efficient Semantic Web Query Processing. Lecture Notes in Computer Science: Advances in Data and Web
Management, 2007. 4505: p. 428-439.
86. Zhang, J., C.K. Chang, and J. Voas. A Uniform Meta-Model for Mediating Formal Electronic Conferences. in the 28th IEEE Annual International Computer Software and Applications Conference (COMPSAC). 2004. Hong Kong, China.
87. Zhang, J., C.K. Chang, and J.-Y. Chung. Mediating Electronic Meetings. in the IEEE 27th Annual International Computer Software and Applications Conference (COMPSAC). 2003.
Dallas, TX, USA.
88. Zhang, J. Extended Collaboration Description Language (X- CODL). in the 10th IEEE International Conference on Enterprise Distributed Object Computing (EDOC). 2006. Hong Kong, China.
89. Zhang, L.-J., J. Zhang, and H. Cai, Services Computing.
2007: Springer.
90. Arsanjani, A., et al., S3: A Service-Oriented Reference
Architecture. IT Professional, 2007. 9(3): p. 10-17.
91. Zhang, L.-J. and J. Zhang, SOA Reference Architecture, in book chapter of "Web Services Research and Practices". 2008, IGI Global: Hershey, PA, USA.
Reference to this paper should be made as follows: R.DEEPAVARTHINI, (200X) ‘BI-CRITERIA SCHEDULING OF SCIENTIFIC GRID WORKFLOWS’
1.R.Deepavarthini has Passed Master of computer
science in Computer Science & Engg, In M.sc
PASSED 85 Percentage marks
2 Assist Prof. V. Udhaya kumar: Has completed
Master of Engg in Computer
Science & Engg. Currently working as Associate Professor at PRIST UNIVERSITY, Puducherry, India. Total teaching experience-10
years. .
IJSER © 2014 http://www.ijser.org