International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1303
ISSN 2229-5518
STRATAGIES FOR DEVELOPING VACCINES FOR HIV/AIDS.
(REVERSE VACCINOLOGY)
KOORAKULA SREELATHA1, SURENDRA BABU KEELA2
Bangalore City College,Department of Biochemistry, Banglore ,India.
.
ABSTRACT: Acquired Immuno Deficiency Syndrome or AIDS was first recognized in the early 1980’s. Although, with the use of antiretroviral medication, the life expectancy of an HIV infected patient can be elongated temporarily, but there is neither a known cure nor a vaccine for AIDS. This paper shows the approaches undertaken in developing a preventive mechanism against HIV by using epitope driven vaccines which target the global variability of HIV. For this purpose computer driven methods are adapted i.e., Insilco methods for increasing the efficiency of vaccines like Epi- Assembler and Vaccine. Use of broadly reactive and neutralizing antibodies also potentially counteracts HIV-1 diversity. HIV-1 Virus like Particles (VLPs) consists of viral gag proteins which self assemble into particular structures analogous in size, morphology to immature HIV-1 particles. As a non-infectious, replication deficient particles, the VLPs are much safer efficient in developing vaccines when compared to traditional vaccines.
INTRODUCTION
—————————— ——————————
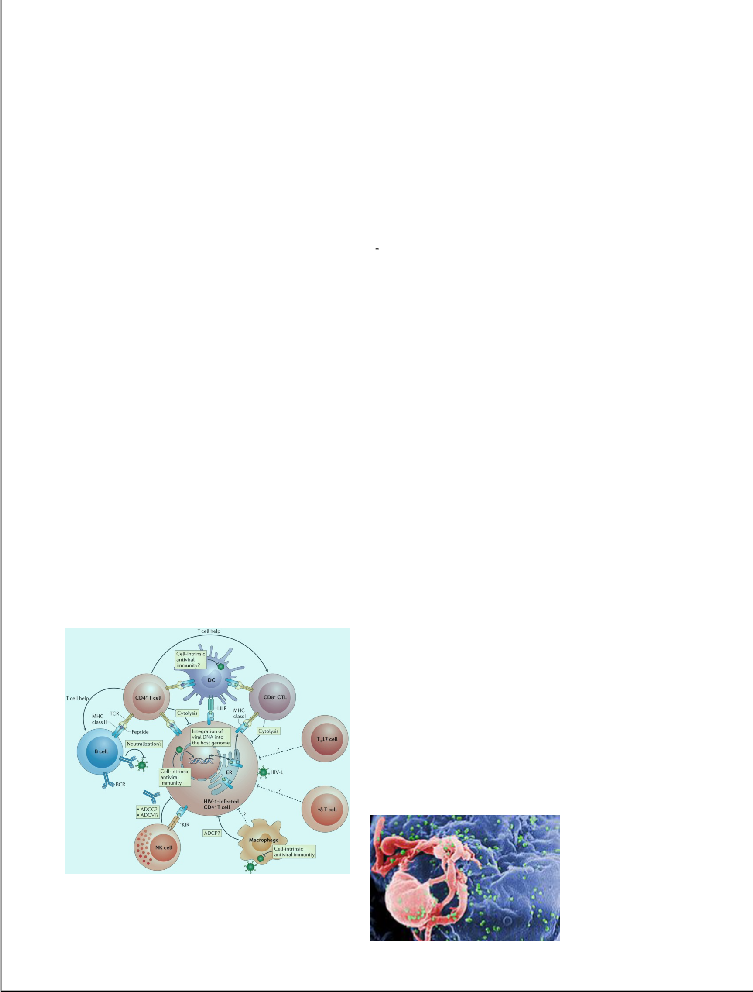
The need for an efficient and an effective defense mechanism against AIDS has never been greater as it is now. Although, with the use of antiretroviral medication, the life expectancy of an HIV infected patient can be
elongated temporarily, but there is neither a known cure nor a vaccine for AIDS. This paper reviews the progress and current approaches undertaken in developing a preventive mechanism for HIV, and thereafter suggests a way to confront the issue by focusing on various vaccine development approaches for HIV, specifically
concentrating on the treatment of HIV-1 (Figure 1).
Fig-1:Replication HIV model
HUMAN IMMUNODEFICIENCY VIRUS
The human immunodeficiency virus (HIV) is a lentivirus (a subgroup of retrovirus) that causes the acquired immunodeficiency syndrome(AIDS), a condition in
humans in which progressive failure of the immune
system allows life-threatening opportunistic infections and cancers to thrive. Without treatment, average survival time after infection with HIV is estimated to be 9 to 11 years, depending on the HIV subtype.Infection with HIV occurs
by the transfer of blood, semen, vaginal fluid, pre-ejaculate, or breast milk. Within these bodily fluids, HIV is present as both free virus particles and virus within infected immune cells.
HIV infects vital cells in the human immune system such as helper T cells (specifically CD4+ T cells), macrophages, and dendritic cells.HIV infection leads to low levels
of CD4+ T cells through a number of mechanisms,
including apoptosis of uninfected bystander cells, direct viral killing of infected cells, and killing of infected
CD4+ T cells by CD8 cytotoxic lymphocytes that recognize
infected cells. When CD4+ T cell numbers decline below a critical level, cell- mediated immunity is lost, and the body becomes progressively more susceptible to opportunistic infections.
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1304
ISSN 2229-5518
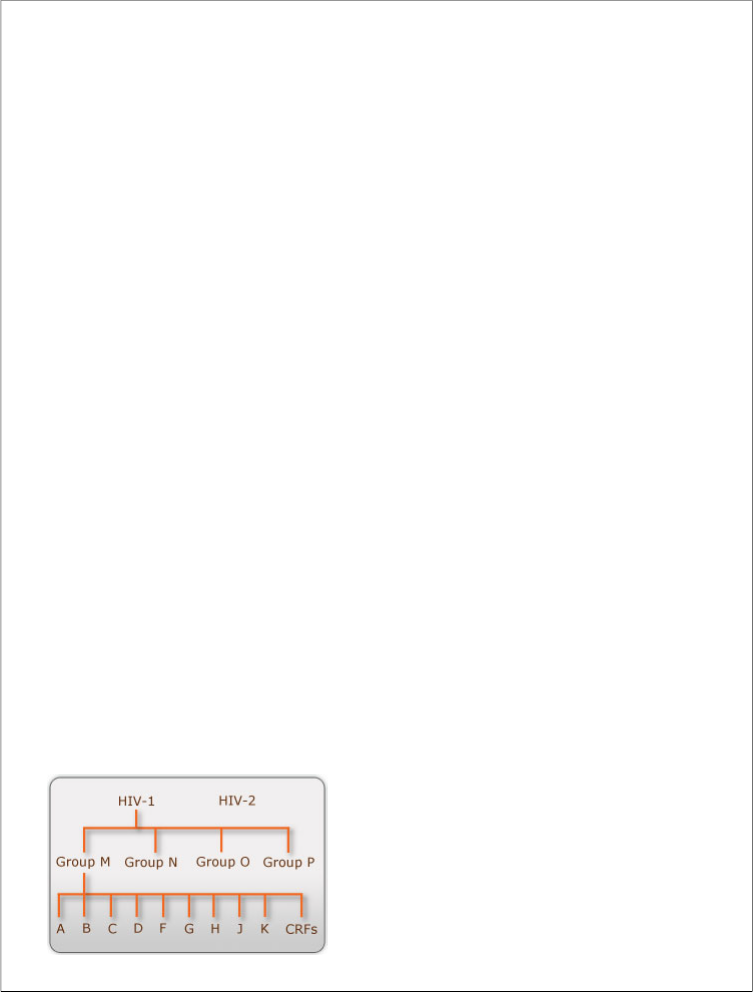
Group: Group VI (ssRNA-RT) Order: Unassigned
Family: Retroviridae Subfamily:Orthoretrovirinae Genus: Lentivirus
Fig:2 HIV binding CD4 cells
Features of HIV-1 that helps in escaping immune detection
Surface Env protein escapes antibody recognition as it possesses variable loops, N-linked glycosylation, conformational flexibility and presence of glycan shield over highly
immunogenic epitopes.
Persistently replicates in the infected individual and keeps on attacking CD 4+ T cells.
Rapidly mutates during infection and thus escapes immune recognition and have extensive viral clade and sequence diversity.
Persist indefinitely as latent proviral DNA, capable of replication in individuals at a later time.
Types of HIV virus
HIV is a highly variable virus which mutates very readily. This means there are many different strains of HIV, even within the body of a single infected person.
Based on genetic similarities, the numerous virus strains may be classified into types, groups and subtypes.
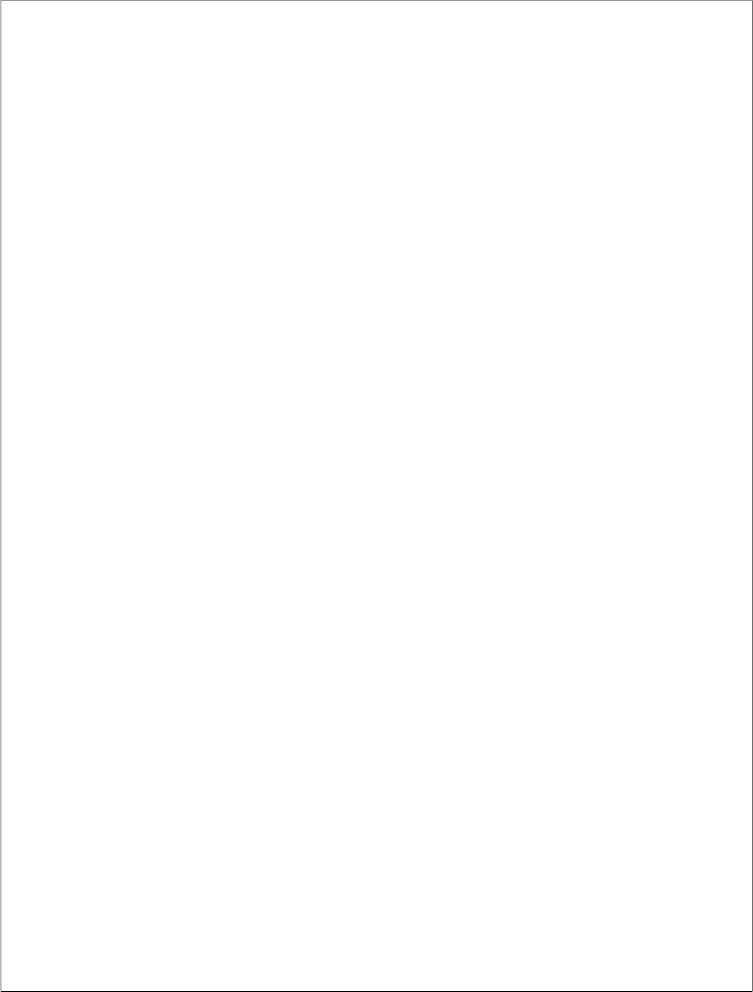
There are two types of HIV: HIV-1 and HIV-2. Both types are transmitted by sexual contact, through blood, and from mother to child, and they appear to cause clinically indistinguishable AIDS. However, it seems that HIV-2 is less easily transmitted, and the period between initial infection and illness is longer in the case of HIV-2.
The strains of HIV-1 can be classified into four groups: the "major" group M, the "outlier" group O and two new groups, N and P. These four groups may represent four separate introductions of simian immunodeficiency virus into humans.
Fig:3HIV types, groups and subtypes
Group O appears to be restricted to west-central Africa and group N - a strain discovered in 1998 in Cameroon - is extremely rare. In 2009 a new strain closely relating to gorilla simian immunodeficiency virus was discovered in a Cameroonian woman. It was designated HIV-1 group
P. 1 More than 90 percent of HIV-1 infections belong to HIV-1 group M and, unless specified, the rest of this page will relate to HIV-1 group M only.
Within group M there are known to be at least nine genetically distinct subtypes (or clades) of HIV-1. These are subtypes A, B, C, D, F, G, H, J and K.
Occasionally, two viruses of different subtypes can meet in the cell of an infected person and mix together their genetic material to create a new hybrid virus (a process similar to sexual reproduction, and sometimes called "viral
sex"). Many of these new strains do not survive for long, but those that infect more than one person are known as "circulating recombinant forms" or CRFs. For example, the CRF A/B is a mixture of subtypes A and B.
The classification of HIV strains into subtypes and CRFs is a complex issue and the definitions are subject to change as new discoveries are made. Some scientists talk about subtypes A1, A2, A3, F1 and F2 instead of A and F, though others regard the former as sub-subtypes
HIV-1 is found to be genetically more diverse than HIV-2 in the envelope region comprising C2, V3 and C3 region. To confirm this further, statistical analysis was performed on HIV-1 and HIV-2 infected patients worldwide. Helena Barroso et al. took HIV-2 sequences from Portugal, belonging to group A, and the majority of HIV-1 sequences belonging to subtype B (75%) followed by subtypes G, C and F.
The time duration of initial infection to final stage in case
of HIV-2 infection is much attenuated as compared to HIV-
1 infection which quickly spreads throughout the host body. This difference in immunological response is expected to be associated with the lower state of immune activation in HIV-2, which may be related to the immunosuppressive activity of the C2-V3-C3 envelope region.
On comparing the amino acid diversity in the C2, V3 and C3 regions of HIV-1 and HIV-2 by Shannon’s entropy studies confirmed that these regions are more variable in HIV-1 than in HIV-2. Entropy results also revealed the fact that HIV-1 possesses high entropy as compared to HIV-2 in each separate region. The region with higher mean
entropy was C3 in both viruses for HIV-1 and also for HIV-
2. V3 region was found to be the least entropic region
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1305
ISSN 2229-5518
both in HIV forms. Amino acids with high entropy were principally found to be present in the C3 region of both viruses and there were more highly entropic amino acids in C3 in HIV-1 than in HIV-2 both in the Portuguese and Control datasets. It has been recently found that HIV-2 displays a faster evolutionary rate in the envelope gp125 and C2-V3-C3 region than HIV-1 in patients who are at an advanced stage of disease .
VACCINES
Vaccines are the medical product designed to activate immune system of the recipient in order to control or prevent infection. In case of HIV-1, mostly preventive vaccines are being developed. These vaccines are designed to protect normal population from getting a HIV infection. Preventive vaccines are widely used to prevent diseases like the flu, chicken pox, measles, mumps, rubella, polio, and hepatitis A and B. Different types of preventive vaccines include subunit vaccines, recombinant vector vaccine, DNA vaccine.
PREVENTIVE HIV-1 VACCINE DESIGN
Due to the escape mechanisms against immune system and unique features of HIV-1, traditional vaccine designs were not of much success. These strategies have proved be successful against many other diseases and pathogens but failed to provide any positive result against HIV-1.
Types of Experimental HIV vaccines based on conventional approaches:
Peptide vaccine
Recombinant subunit protein vaccine
Live vector vaccine
DNA vaccine
Recent Advances
Due to the absence of a vaccine that can prevent HIV infection,stringent research was started to develop a vaccine that delays or prevents the progression of HIV infection which could be an important intervention to curtail the current global epidemic of HIV. Recently large scale clinical study- RV144 showed some modest response (31%) in reduction of HIV-1 infection [34]. RV144 vaccine consisted of live recombinant viral vector ALVAC-HIV-1, consisting proteins gag/pol/env as prime vaccine
followed by booster injections carrying gp120 region
(AIDSVAX B/E) as protein boost (Table 3). More than 16,000 people were subjected to trial of RV144 in Thailand. This vaccine targeted both humoral and cell mediated immune response and vaccine
recipients exhibited antibody dependent cell mediated cytotoxicity responses . Another large scale clinical study, HVTN 505, also targeted the humoral and cellular immune responses. It consisted of multiclade HIV-1 DNA plasmid (EnvA, EnvB, EnvC, gagB, polB, nefB) followed by booster dose of recombinant adenovirus vector (Ad5
EnvA, EnvB, EnvC, gag/polB).
HIV VACCINE DEVELOPMENT BY COMPUTER AIDED DESIGN
In the year 2005, scientist came up with the approach of computer aided drug designing against HIV-1. They introduced the concept of designing epitope driven vaccines that target the global variability of HIV. The
scientists developed computer driven methods for speeding up the process of vaccine development. These methods include the use of Epi-Assembler which derives “Immunogenic Consensus Sequence” (ICS) epitopes from multiple viral variants, and Vaccine CAD, which reduces junctional immunogenicity when epitopes are aligned in a string-of-beads format for insertion into a DNA expression vector.
In this research 20 consensus sequences were isolated from HIV-1 peptides that were found to be immunogenic. There immunogenicity was confirmed computationally by the Epi
- Assembler. The core 9-mer contained in these consensus
peptides was conserved in approximately 105 to 2250 individual HIV-1 strains. Nineteen of the twenty ICS epitopes (95%) evaluated in this study was confirmed in ELISpot assays using peripheral blood monocytes obtained from 13 healthy HIV-1 infected subjects. Twenty five ICS peptides (all 20 of the peptides evaluated in this study and 5 additional ICS epitopes) were then aligned in a pseudo protein string using “VaccineCAD”, an epitope alignment tool that eliminates immunogenicity created by the
junctions between the epitopes. They reordered the
construct to reduce the immunogenicity of the junctions between epitopes as measured by EpiMatrix, an epitope mapping algorithm. The reordered construct was also a more effective immunogen in vivo when tested in HLA-DR transgenic mice.
GENETIC DIVERSITY CONSIDERATIONS FOR VACCINE DESIGN
The genetic diversity of HIV-1 across the globe is a major challenge for developing an HIV vaccine. To facilitate immunogen design, it is important to characterize clusters of commonly targeted T-cell epitopes across different HIV
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1306
ISSN 2229-5518
clades. For examining the conserved sequences present in cross clades, a team of scientist examined 39 HIV-1 clade C infected individuals for IFN-γ Gag-specific T-cell responses using five sets of overlapping peptides, two sets matching clade C vaccine candidates derived from trains from South Africa and China, and three peptide sets corresponding to consensus clades A, B, and D sequences . Previous results have shown that HIV-specific T-cells are cross-reactive among different HIV clades but with a preference for the infecting clade.The results from these studies demonstrated that within a single individual, some HIV peptides were exclusively recognized in the clade C sequence variants (CDu422 and CCH), whilst others were uniquely recognized in the clades B, A and D peptide variant. The recognition of clades B, A and D peptide
variants and not the corresponding clade C peptide variants is of importance, as it demonstrates that using a single peptide reagent set leads to a considerable number of responses being missed when investigating T-cell immune responses.
These findings are of interest, since a previous study demonstrated no increase in epitope recognition when using the center of the tree(COT) and most common recent ancestor (MRCA) peptide sequences in addition to clade B consensus peptides, in a clade B HIV-infected population.
VIRUS LIKE PARTICLES (VLPS)
Recently a major approach came into light which focused to exploit the conserved regions of IV-1 proteins by overcoming viral immune evasion. gp120 consists of combination of conserved and variable domains. It have five conserved domain namely C1- C5 which are separated by variable domains- V1- V5 . The envelope(transmembrane) protein gp120 co-receptor
inding site can also actas target. Inducing production of broadly reactive neutralizing antibodies could also potentially counteract HIV-1 diversity. Many antibodies like PG9, PG16 target even the conserved subdomain
within the V1/V2 region. PGT 121- PGT123 and PGT 125-
131 are potent enough to recognize the conserved base of V3 domain and interestingly antibodies against these regions are the one to be elicited first in HIV-1 infection. These potential solutions for HIV-1 vaccine development could be potentially realized by exploiting prophylactic HIV-1 virus-like particle (VLP) vaccines.
FEATURES OF VLP’S
VLPs express multiple viral epitopes that stimulate a diverse set of immune responses, without many of the deleterious effects of a live-attenuated virus
Potential for activating
both endogenous and exogenous antigens
processing pathways, leading to presentation of viral peptides by MHC class I and class II molecules
VLPs may be more cost efficient than co-inoculating multiple single gene vaccines for future Phase I clinical trials
T Cell based vaccine
The previous vaccine attempts mainly focused on eliciting antibody response in patients, but now due to the failures or inefficiency of this approach, focus has been shifted to T cell based vaccines. These vaccines are expected to control virus replication rather than preventing infection because they target infected cells by recognizing the viral proteins presented on major histocompatibility complex (MHC) Proteins . Thus the main aim of these vaccines is to control viremia. A T cell based vaccine consists of epitopes of surface proteins and inside the virion. These DNA vaccines work by introducing the viral antigens to T cells through MHC complex class I and class II. The immunogenicity of these vaccines can be enhanced by new DNA delivery methods . Poxvirus, adenovirus and cytomegalovirus
vectors are some of the vectors that can be used in T cell based vaccine Other than these replication incompetent adenoviruses have also been used that expresses HIV-1 immunogens and elicits CD4+ and CD8+ T cells response in recipient individual.
CONCLUSION
The basic idea behind this work is to develope effective potential vaccines which can fight against Human Immune Virus( HIV). The immune system employs mature defense mechanism which includes antibodies and numerous phagocytic cells. Early vaccine researches has mainly designed on the immune system competency by producing antibodies which would block HIV infection. However, such ideas failed in clinical trials because antibodies worked only against lab-cultured HIV, but not on wild strains of the HIV. As the results are not prooving enough and in need for the development of more sophisticated and quicker techniques regarding the same a promising remedy in this area is the use of reverse vaccinology approaches which includes various computational approaches for epitope prediction and their binding efficiency levels with broadly neutralizing antibodies. Following the prediction of a possible vaccine by in silico methods, their efficacy can
be evaluated by IFN-γ ELISPOT assay,intracellular
cytokine staining assay, virus neutralization assay, ELISA binding antibodies. Use of reverse vaccinology and immunoformatics in silico approaches will help in speeding up time for vaccine development which is cost effective.
ACKNOWLEDGEMENT:
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1307
ISSN 2229-5518

I Sincerely thank
MICELLES LIFESCIENCES PRIVATE LIMITED
, Lucknow for providing technical details and guidance .RFERENCES
1.Grossman Z, Meier-Schellersheim M, Sousa AE, Victorino RM,
Paul W E (2002) CD4+ T-cell depletion in HIV infection: are we closer to understanding the cause? Nat Med 8: 319-323.
2. Korber BT, Kunstman KJ, Patterson BK, Furtado M, McEvilly MM, et al. (1994) Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. Journal of Virology 68:
7467-7481.
3. Borrego P, Marcelino JM, Rocha C, Doroana M, Antunes F, et al. (2008) The role of the humoral immune response in the molecular evolution of the envelope C2, V3 and C3 regions in chronically HIV-2 infected patients. Retrovirology 5: 78.
4. Skar H, Borrego P, W allstrom TC, Mild M, Marcelino JM, et al. (2010) HIV- 2 genetic evolution in patients with advanced disease is faster than that in matched HIV-1 patients. J Virol 84: 7412-7415.
5. National Institute of Allergy and Infectious Diseases. http://www.pnac.net.pk/Reports/FactSheets/English/FactSheet-1.pdf.
6. Peters BS, Conway K (2011) Therapy for HIV: past, present, and future. Adv Dent Res 23: 23-27.
7. Fultz PN (1997) AIDS and Other Manifestations of HIV Infection. (3rd edn), Newyork 201-215.
8. Girard M, Meignier B, Barré-Sinoussi F, Kieny MP, Matthews T, et al. (1995) Vaccine-induced protection of chimpanzees against infection by a heterologous human immunodeficiency virus type 1. J Virol 69: 6239-6248.
9. Letvin NL, King NW (1990) Immunologic and pathologic manifestations of the infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Acquir Immune Defic Syndr
3: 1023-1040.
10. Myers G, Korber B, Berzofsky JA, Pavlakis GN, Smith RE (1993) A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. Human Retroviruses and AIDS Eds.
11. Igarashi T, Shibata R, Hasebe F, Ami Y, Shinohara K, et al.
(1994) Persistent infection with SIVmac chimeric virus having tat, rev,
vpu, env and nef of HIV type 1 in macaque monkeys. AIDS Res Hum
Retroviruses 10: 1021-1029.
12. Luciw PA, Pratt-Lowe E, Shaw KE, Levy JA, Cheng-Mayer C (1995) Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc Natl Acad Sci U S A 92: 7490-7494.
13. Li JT, Halloran M, Lord CI, W atson A, Ranchalis J, et al. (1995) Persistent infection of macaques with simian-human immunodeficiency viruses. J Virol 69: 7061-7067.
14. Li J, Lord CI, Haseltine W , Letvin NL, Sodroski J (1992) Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus that expresses the HIV-1 envelope glycoproteins. J Acquir Immune Defic Syndr 5: 639-646.
15. Reimann KA, Li JT, Veazey R, Halloran M, Park IW , et al. (1996) A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol 70: 6922-6928.
16. Nishimura Y, Igarashi T, Donau OK, Buckler-W hite A, Buckler C, et al. (2004) Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc Natl Acad Sci U S A 101: 12324-12329.
17. Williams R, Bokhari S, Silverstein P, Pinson D, Kumar A, et al. (2008) Nonhuman primate models of NeuroAIDS. J Neurovirol 14:
292-300.
18. van Baalen CA, Pontesilli O, Huisman RC, Geretti AM, Klein
MR,
de W olf F, Miedema F, Gruters RA, Osterhaus AD: Human immunodeficiency virus type 1 Rev- and Tat-specific cytotoxic T lymphocyte frequencies inversely correlate with rapid progression to AIDS. J Gen Virol 1997, 78:1913-1918.
19. Venet A, Bourgault I, Aubertin AM, Kieny MP, Levy JP: Cytotoxic T
lymphocyte response against multiple simian immunodeficiency virusA (SIV) proteins in SIV-infected macaques. J Immunol 1992, 148:2899-2908.
20.
Butto S, Fiorelli V, Tripiciano A, Ruiz-Alvarez MJ, Scoglio A, Ensoli F, Ciccozzi M, Collacchi B, Sabbatucci M, Cafaro A et al.: Sequence conservation and antibody cross-recognition of
clade B human immunodeficiency virus (HIV) type 1 Tat protein in HIV-1-infected Italians, Ugandans, and South Africans.
J Infect Dis 2003, 188:1171-1180.
Demonstrates conservation and cross-recognition of immunodominant
B-cell epitopes across different HIV-1 clades (A to D).
21.
_
Bobbitt KR, Addo MM, Altfeld M, Filzen T, Onafuwa AA,
W alker BD, Collins KL: Rev activity determines sensitivity of
HIV-1-infected primary T cells to CTL killing. Immunity 2003,
18:289-299.
Reports a novel mechanism by which HIV can escape immune recognition.
22. Addo MM, Altfeld M, Rosenberg ES, Eldridge RL, Philips
MN,
Habeeb K, Khatri A, Brander C, Robbins GK, Mazzara GP et al.: The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from
HIV-1-infected individuals. Proc Natl Acad Sci USA 2001,
98:1781-1786.
23.
_
de Oliveira T, Salemi M, Gordon M, Vandamme AM,
van Rensburg EJ, Engelbrecht S, Coovadia HM, Cassol S: Mapping sites of positive selection and amino acid diversification in the HIV genome: an alternative approach to vaccine design? Genetics 2004, 167:1047-1058.
An interesting approach to define evolutionary patterns that may help
identify relevant HIV-1 epitopes and vaccine design.
24. Fackler OT, Baur AS: Live and let die: Nef functions beyond
HIV
replication. Immunity 2002, 16:493-497.
25. Kestler HW III, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC: Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 1991, 65:651-662.
26. Collins KL, Chen BK, Kalams SA, W alker BD, Baltimore D: HIV-1
Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 1998, 391:397-401.
27. Swingler S, Mann A, Jacque J, Brichacek B, Sasseville VG, Williams K, Lackner AA, Janoff EN, W ang R, Fisher D, Stevenson M: HIV-1 Nef mediates lymphocyte chemotaxis
and activation by infected macrophages. Nat Med 1999,
5:997-1003.
28. Swingler S, Brichacek B, Jacque JM, Ulich C, Zhou J, Stevenson M: HIV-1 Nef intersects the macrophage CD40L signalling pathway to promote resting-cell infection.
Nature 2003, 424:213-219.
29.
_
Sol-Foulon N, Moris A, Nobile C, Boccaccio C, Engering A,
Abastado JP, Heard JM, van Kooyk Y, Schwartz O: HIV-1
Nefinduced
upregulation of DC-SIGN in dendritic cells promotes lymphocyte clustering and viral spread. Immunity 2002,
16:145-155.
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1308
ISSN 2229-5518
Describes a novel immunodysregulatory mechanism by which
Nef can
contribute to HIV-1 virulence and pathogenesis.
30. Addo MM, Yu XG, Rosenberg ES, W alker BD, Altfeld M: Cytotoxic
T-lymphocyte (CTL) responses directed against regulatory
and accessory proteins in HIV-1 infection. DNA Cell Biol 2002,
21:671-678.
IJSER © 2014 http://www.ijser.org





