The subject of current investigation are three of MpGA
ty/non-validity can be obtained as follows:
modifications with the slectIion opeJrator on firstSplace, namely ER
MpGA-SCM, MpGA-SMC and MpGA-SC.
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1870
ISSN 2229-5518
Quality Assessment of Multi-population
Genetic Algorithms Performance
Tania Pencheva, Maria Angelova, Krassimir Atanassov
Abstract— The quality of performance of multi-population genetic algorithms (MpGA) has been assessed for the purposes of parameter identification of S. cerevisiae fed-batch cultivation. Intuitionistic fuzzy logic has been implemented aiming to derive intuitionistic fuzzy estimations of obtained model parameters. Three kinds of MpGA, differ from each other in the sequence of execution of main genetic operators, namely selection, crossover and mutation, have been assessed before and after the application of the recently developed procedure for purposeful model parameters genesis. Results obtained after the implementation of intuitionistic fuzzy logic for MpGA performances assessment have been compared and MpGA with a sequence selection and crossover after the procedure for purposeful model parameters genesis application has been distinguished as the fastest and quite reliable one.
Index Terms— Quality assessment, intuitionistic fuzzy logic, multi-population genetic algorithms, parameter identification, yeast fed-batch cultivation.
—————————— ——————————
Genetic algorithms (GA) [10] are one of the most representa- tive examples of methods based on biological evolution. GA are inspired by Darwin’s theory of “survival of the fittest” and they are based on the mechanics of natural selection and ge- netics. GA seek for the global optimal solution in complex multidimensional search space by simultaneously evaluating
1. [Start]
Generate k random subpopulations each of them with n
chromosomes
2. [Objective function]
Evaluate the objective function of each chromosome x
in the subpopulation
IJSER
many points in the parameter space. Their properties such as
hard problems solving, noise tolerance, easiness to interface
and hybridize, make GA a suitable and quite workable tool
especially for tasks which are not completely determined.
Such a real challenge for researchers is the parameter identifi- cation of fermentation processes [1], [2], [12], [13], [14] which modeling is a specific task, rather difficult to be solved. The idea genetic algorithms to be tested as an alternative technique
for parameters identification of fermentation process model has been provoked by the inability of conventional optimiza- tion methods to reach to a satisfactory solution [2], [13].
Goldberg [10] initially presents the standard single- population genetic algorithm (SGA) inspired by natural genet- ics, which searches for a global optimal solution using three main genetic operators in a sequence selection, crossover and mutation. More similar to nature is multi-population genetic algorithm (MpGA), since there many populations, called sub- populations, evolve independently from each other. After a certain number of generations (isolation time), a part of indi- viduals are distributed between the subpopulations (migra- tion).
According to [10], [11], working principle of standard
MpGA is shown in the following nine-step procedure:
————————————————
Institute of Biophysics and Biomedical Engineering
Bulgarian Academy of Sciences
105 Acad. Georgi Bonchev Str., 1113 Sofia, Bulgaria E-mails: tania.pencheva@biomed.bas.bg, maria.angelova@biomed.bas.bg, krat@bas.bg
3. [Fitness function]
Find the fitness function of each chromosome x in the
subpopulation
4. [New population]
Create a new population by repeating following steps:
4.1. [Selection]
Select parent chromosomes from the subpopula-
tion according to their fitness function
4.2. [Crossover]
Cross over the parents to form new offspring with
a crossover probability
4.3. [Mutation]
Mutate new offspring at each locus with a muta-
tion probability
5. [Accepting]
Place new offspring in a new population
6. [Replace]
Use new generated population for a further run of the
algorithm
7. [Migration]
Migration of individuals between the subpopulations
after following isolation time
8. [Test]
If the end condition is satisfied, stop and return the best
solution in current population, else move to Loop step
9. [Loop]
Go to Fitness step.
Since the basic idea of GA is to imitate the mechanics of natural selection and genetics, one can make an analogy with the processes occurring in nature, saying that the probability one of the genetic operators to be executed before or after the other is comparable to the probability that processes to occur in a reverse order. Thus, following that idea, altogether six
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1871
ISSN 2229-5518
modifications of MpGA have been developed and investigated towards model accuracy and algorithms convergence time. All considered MpGA have a similar structure and follow the nin- step procedure presented above, but differ from each other in the sequence of execution of the main genetic operators selec- tion, crossover and mutation. The standard MpGA, originally presented in [10], is here denoted as MpGA_SCM (coming from sequence selection, crossover, mutation). MpGA_CMS (crossover, mutation, selection), MpGA_SMC (selection, muta- tion, crossover) and MpGA_MCS (mutation, crossover, selec- tion) have been proposed in [6]. Another two kinds of MpGA, omitting mutation operator, namely MpGA-SC (selection, crossover) and MpGA_CS (crossover, selection), are further developed [4] provoked by promising results obtained in SGA [3].
The sequence of algortihm steps in different kinds of MpGA, according to the presented above nine-step procedure, is as follows:
rithms, a great number of algorithm runs have to be executed if one would like to obtain reliable results no matter of the object considered. Firstly, the genetic algorithm searches for solutions of model parameters in wide but reasonably chosen boundaries according to the statements in [14] – so-called “broad” range. When results from many algorithms execu- tions were accumulated and analyzed, they showed that the values of model parameters can be assembled and predefined boundaries of model parameters could be restricted. This is the main idea of PMPG, which results in the defining of more appropriate boundaries for variation of the model parameters values. The procedure application leads to decrease conver- gence time while at least saving or even improving the model accuracy.
In intuitionistic fuzzy logic (IFL) [7], [8] if p is a variable then its truth-value is represented by the ordered couple
The subject of current investigation are three of MpGA
ty/non-validity can be obtained as follows:
modifications with the slectIion opeJrator on firstSplace, namely ER![]()
MpGA-SCM, MpGA-SMC and MpGA-SC.
M ( p )
![]()
= m , N ( p )
= 1 - n , (2)
The evaluation of the quality of any algorithm performance could be based on some representative criteria such as the ob- jective function value and the algorithm convergence time. As an alternative for assessing the quality of different algorithms intuitionistic fuzzy logic (IFL) might be applied for various purposes. IFL is based on the construction of degrees of validi- ty and non-validity which requires the algorithms to be per- formed in two different intervals of model parameters varia- tion. One interval could be so-called “broad” range known
u u
where m is the lower boundary of the “narrow” range; u – the upper boundary of the “broad” range; n – the upper boundary of the “narrow” range.
If there is a database collected having elements with the form <p, M(p), N(p)>, different new values for the variables can be obtained. In case of considered here three records in the database, the following new values can be obtained:
Vstrong _ opt = < M1 ( p ) + M 2 ( p ) + M 3 ( p ) –
from the literature [14]. The other one, called “narrow” range, is user-defined and might be obtained using different criteria – e.g. based on the minimum and maximum values, or on the average ones, or after the implementation of the recently de-
− M1 ( p ) M 2 ( p ) – M1 ( p ) M 3 ( p ) – M 2 ( p ) M 3 ( p ) +
+ M1 ( p ) M 2 ( p ) M 3 ( p ), N1 ( p ) N2 ( p ) N3 ( p ) >
Vopt = < max (M1 ( p ), M 2 ( p ), M 3 ( p )),
(3)
veloped procedure for purposeful model parameters genesis
[5].
min ( N1 ( p ), N2 ( p ), N3 ( p )) >
, (4)
The aim of this study is intuitionistic fuzzy estimations to V
= < (M
( p ) + M
( p ) + M
( p )) / 3,
be applied for assessing the quality of performance of three
different kinds of MpGA, namely MpGA-SCM, MpGA-SMC
and MpGA-SC, for the purposes of parameter identification of
aver
1 2 3
( N1 ( p ) + N2 ( p ) + N3 ( p )) / 3) >
, (5)
S. cerevisie fed-batch cultivation. Aiming to save decreased convergence time while keeping or even improving model accuracy, intuitionistic fuzzy estimations overbuild the results obtained after procedure of purposeful model parameters genesis.
Vpes = < min (M1 ( p ), M 2 ( p ), M 3 ( p )),
max ( N1 ( p ), N2 ( p ), N3 ( p )) >
Vstrong _ pes = < M1 ( p ) M 2 ( p ) M 3 ( p ),
N1 ( p ) + N2 ( p ) + N3 ( p ) −
, (6)
(7)
The procedure for purposeful model parameter genesis (PMPG) has been originally developed in [5] and firstly ap- plied for parameter identification of S. cerevisiae using simple genetic algorithms. Due to the stochastic nature genetic algo-
– N1 ( p ) N2 ( p ) – N1 ( p ) N3 ( p ) – N2 ( p ) N3 ( p ) +
+ N1 ( p ) N2 ( p ) N3 ( p ) >
Therefore, for each p
Vstrong _ pes ( p ) ≤ Vpes ( p ) ≤ Vaver ( p ) ≤ Vopt ( p ) ≤ Vstrong _ opt ( p ) .
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1872
ISSN 2229-5518
feeding solution, [g/l]; μ , q S , q E ,
q – specific
2
As mentioned above, the implementation of IFL for assess- ment of genetic algorithms performance quality requires con- struction of degrees of validity and non-validity in two differ- ent intervals of model parameters variation: so-called “broad” range as known from the literature and so-called “narrow” range which is user-defined. Here the authors applied a pro- cedure for assessment of algorithm quality performance (AAQP) implementing IFL. The procedure starts with perfor- mance of a number of runs of each of the algorithms, object of
growth/utilization rates of biomass, substrate, ethanol and
dissolved oxygen, [1/h]. All functions are continuous and dif-
ferentiable.
The fed-batch cultivation of S. cerevisiae considered here is
characterized by keeping glucose concentration equal to or
below its critical level (Scrit = 0.05 g/l), sufficient dissolved oxygen O2 ≥ O2crit (O2crit = 18%) and availability of ethanol in
the broth. This state corresponds to the so called mixed oxida-
tive state (FS II) according to functional state modeling ap-
proach [13]. Hence, specific rates in Eqs. (8)-(12) are:
the investigation in both “broad” and “narrow” ranges of model parameters. Then the average values of the objective
μ= μ2 S
S
![]()
S +kS
+ μ2 E
![]()
E ,
E +kE
μ
![]()
q = 2 S
SX
S ,
S +kS
function, algorithms convergence time and each of the model
parameters for each one of the ranges and each one of the in-
vestigated algorithms are obtained. According to (2), degrees![]()
q = - μ2 E E ,
E Y E +k
qO = qE YOE + qSYOS , (13)
of validity/non-validity for each of the algorithms, object of
where
μ2 S , μ2 E
are the maximum growth rates of substrate
the investigation, are determined. Then, in case of three ob- jects, strong optimistic, optimistic, average, pessimistic and strong pessimistic values are calculated for each one of the model parameters according to (3)-(7). Next determined in such way values are assigned to each of the model parameters
for each of the ranges for each of the algorithms. Finally, based
and ethanol, [1/h]; kS , kE – saturation constants of substrate
and ethanol, [g/l]; Yij – yield coefficients, [g/g]; and all model parameters fulfill the non-zero division requirement.
As an optimization criterion, mean square deviation be- tween the model output and the experimental data obtained during cultivation has been used:
on these assigns, the quality of each one of considered algo-
J = ∑(Y - Y * )2 → min ,
(14)
rithm is assessed.
IJSEwhere Y is the eRxperimental data, Y* – model predicted data,
Experimental data of S. cerevisiae fed-batch cultivation is ob- tained in the Institute of Technical Chemistry – University of Hannover, Germany [13]. The cultivation of the yeast S. cere- visiae is performed in a 2 l reactor, using a Schatzmann medi- um. Glucose in feeding solution is 35 g/l. The temperature was controlled at 30°C, the pH at 5.5. The stirrer speed was set to 1200 rpm. Biomass and ethanol were measured off-line, while substrate (glucose) and dissolved oxygen were meas- ured on-line.
Mathematical model of S. cerevisiae fed-batch cultivation is commonly described as follows, according to the mass balance [13]:
Y = [X, S, E, O2 ].
The procedure for purposeful model genesis has been applied to parameter identification of S. cerevisiae fed-batch cultivation using three kinds of MpGA. Following model (8)-(13) of S. cerevisiae fed-batch cultivation, nine model parameters have been estimated altogether, applying consequently MpGA_SCM, MpGA_SMC and MpGA_SC. The values of GA parameters and type of genetic operators in MpGA considered here are tuned according to [6]. GA is terminated when a cer- tain number of generations is fulfilled, in this case 100. Scalar relative error tolerance RelTol is set to 1e-4, while the vector of absolute error tolerances (all components) AbsTol – to 1e-5. Pa-![]()
![]()
dX =μX - F X
(8)
rameter identification of the model (8)-(12) has been per-
dt V
![]()
![]()
dS = -q X + F
S - S
(9)
formed using Genetic Algorithm Toolbox [9] in Matlab 7 envi-
ronment. All the computations are performed using a PC Intel
dt S
V ( in )
Pentium 4 (2.4 GHz) platform running Windows XP.
The quality of MpGA performance is assessed before and![]()
![]()
dE = q X - F E
(10)
after application of PMPG, that means that the “narrow” range
dt E V
is obtained applying PMPG. The obtained results are firstly
dO2 = -q X + k O2 a (O * - O )
(11)
analyzed according to achieved objective function values and![]()
dt O2
L 2 2
convergence time. For each of MpGA investigated here the
minimum and the maximum of the objective function are de-![]()
dV =F , (12)
dt
where X is the concentration of biomass, [g/l]; S – concentra- tion of substrate (glucose), [g/l]; E – concentration of ethanol,
termined, and the levels of performance are constructed ac- cording to PMPG [5]. For each of the levels, obtained in such a way, the minimum, maximum and average values of each model parameter have been determined. The new boundaries
[g/l]; O2 – concentration of oxygen, [%]; O*
– dissolved oxy-
of the model parameters are constructed in a way that the new
gen saturation concentration, [%]; F – feeding rate, [l/h];
minimum is lower but close to the minimum of the top level,
V – volume of bioreactor, [l];
k O2 a
– volumetric oxygen trans-
and the new maximum is higher but close to the maximum of
fer coefficient, [1/h]; Sin – initial glucose concentration in the
the top level. Table 1 presents previously used “broad”
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1873
ISSN 2229-5518
boundaries for each model parameter according to [14] as well as new boundaries proposed based on PMPG when applying
MpGA. Additionally, Table 1 consists of intuitionistic fuzzy estimations, obtained based on (2).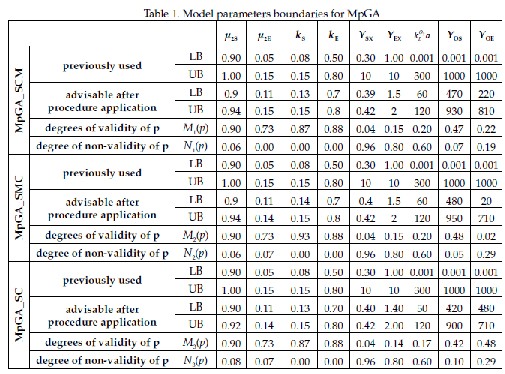
IJSER
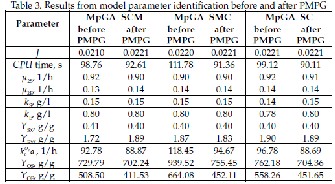
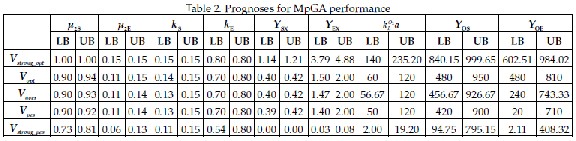
Table 2 presents the boundaries (low LB and up UB) for the strong optimistic, optimistic, average, pessimistic and strong pessimistic prognoses for the performances of MpGA algorithm, obtained based on intuitionistic fuzzy estimations (2) and formula (3)-(7).
Investigated here three kinds of MpGA have been again applied for parameter identification of S. cerevisiae fed-batch cultivation involving newly proposed according to Table 1 boundaries. Several runs have been performed in order reliable results to be obtained. Table 3 presents the average values of the objective function, convergence time and model parameters when MpGA_SCM, MpGA_SMC and MpGA_SC have been executed in “broad” and “narrow” ranges.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1874
ISSN 2229-5518
It is worth to note that in three considered here kinds of MpGA, running of algorithms in “narrow” range leads to ex- pecting decrease of the convergence time while saving the high model accuracy, as it was shown in the case of SGA and also expected here. Running MpGA in “narrow” range reduc- es the computation time 1.07, 1.22 and 1.10 times when MpGA_SCM, MpGA_SMC and MpGA_SC have been respec- tively applied. In addition the results obtained in the “narrow” range hit the top level of performance, thus showing good effectiveness of all considered here kinds of MpGA.
Table 4 lists the number and type of the estimations as- signed to each of the parameters for three considered here kinds of MpGA, applied and before and after the PMPG.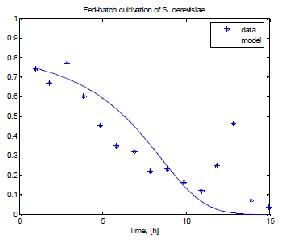
 a) biomass concentration
a) biomass concentration
As seen from Table 4, there are no any strong pessimistic and pessimistic prognoses. One of the cases significantly stands out of the others with 4 strong optimistic and 5 optimis- tic prognoses – this is the case of MpGA_SMC in “broad” range. Moreover, this is the case with the highest model accu- racy achieved, but in the same time this is the slowest one case among all considered here. It is quite interesting that all MpGA in “narrow” range have been assessed absolutely equally – with 2 strong optimistic, followed by 6 optimistic and 1 average prognoses. In these three distinguished as more reliable cases, the value of the objective function is almost
equal to the lowest one that means they are with the highest achieved degree of accuracy. Among them the fastest one is the case of MpGA_SC. Thus, based on the intuitionistic fuzzy estimations of the model parameters and further constructed prognoses, MpGA_SC in “narrow” range is distinguished as an acceptable compromise – not the highly assessed, but the fastest one, if one would like to obtained results with a high level of relevance and for less computational time.
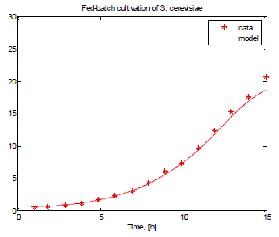
Fig. 1 shows results from experimental data and model prediction, respectively, for biomass, ethanol, substrate and dissolved oxygen when MpGA_SC after the procedure for the purposeful model parameter genesis has been applied.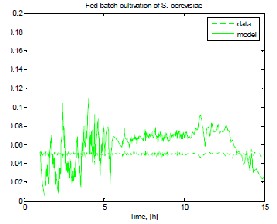
b) ethanol concentration
c) substrate concentration
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1875
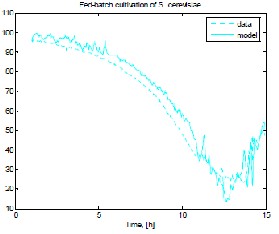
ISSN 2229-5518
d) dissolved oxygen concentration
Fig. 1 Model prediction compared to experimental data
when MpGA_SC in “narrow” range has been applied
Overall, obtained results show the workability and the ef- fectiveness of the procedure for purposeful model parameter genesis.
timization algorithms as well as to be applied to various ob- jects of parameter identification.
This work is partially supported by National Science Fund of
Bulgaria, grant DMU 03-38.
[1] J. Adeyemo, and A. Enitian, “Optimization of Fermentation Processes Using Evolutionary Algorithms – A Review”, Scient. Res. and Ess., vol. 6, no. 7, pp. 1464-1472, 2011.
[2] M. Angelova, S. Tzonkov, and T. Pencheva, “Genetic Algorithms based Parameter Identification of Yeast Fed-batch Cultivation”, Lect. Notes Comp. Sci., vol. 6046, pp. 224-231, 2011.
[3] M. Angelova, and T. Pencheva, “Algorithms Improving Convergence Time in Parameter Identification of Fed-Batch Cultivation”, Comptes rendus de l'Académie bulgare des Sciences, vol.
65, no. 3, pp. 299-306, 2012.
[4] M. Angelova, and T. Pencheva, “Improvement of Multi- population Genetic Algorithms Convergence Time”, Monte Carlo Methods and Application, pp. 1-9, 2013.
[5] M. Angelova, and T. Pencheva, “Purposeful Model Parameters Genesis in Simple Genetic Algorithms”, Computers and Mathematics with Applications, vol. 64, pp. 221-228, 2012.
[6] Angelova M., and T. Pencheva, “Tuning Genetic Algorithm
Parameters to Improve Convergence Time”, International Journal of
Chemical Engineering, Article ID 646917, 2011,
![]()
Intuitionistic fuzzy logic has been implemented in this inves- tigation in order to assess the genetic algorithms quality per- formance for the purposes of parameter identification of S. cerevisiae fed-batch cultivation. Aiming at keeping obtained promising results, namely less convergence time at saved and even improved model accuracy, intuitionistic fuzzy logic overbuilds the results from the application of recently devel- oped procedure for purposeful model parameter genesis. This procedure has been here applied to three kinds of MpGA dif- fer from each other in the sequence of execution of main ge- netic operators. Intuitionistic fuzzy logic has been implement- ed to obtain intuitionistic fuzzy estimations of model parame- ters and further to construct strong optimistic, optimistic, av- erage, pessimistic and strong pessimistic prognoses for the algorithms performances. Thus, based on the intuitionistic fuzzy estimations of the model parameters and further con- structed prognoses, the following summaries can be outlined:
1) the highly assessed MpGA_SMC in “broad” range is the slowest one, thus getting the user to make a decision between reliability and speed; 2) among the three equal performances of MpGA in “narrow” range, MpGA with a sequence selection and crossover and omitting the mutation is the fastest one. As an acceptable compromise, MpGA_SC in “narrow” range is distinguished as not the highly assessed, but the fastest one, if one would like to obtained results with a high level of rele- vance and for less computational time.
Presented here IFL based “cross-evaluation” of three kinds of MpGA demonstrates the workability of intuitionistic fuzzy estimations to assist in assessment of quality of algorithms performance. The estimations based on intuitionistic fuzzy logic might be considered as an appropriate tool for reliable assessment for genetic algorithm parameters, for different op-
http://www.hindawi.com/journals/ijce/2011/646917/
[7] K. Atanassov, “Intuitionistic Fuzzy Sets“, Springer, Heidelberg,
1999.
[8] K. Atanassov, “On Intuitionistic Fuzzy Sets Theory”, Springer, Berlin, 2012.
[9] A.J. Chipperfield, P. Fleming, H. Pohlheim, and C.M. Fonseca, “Genetic Algorithm Toolbox for Use with MATLAB”, User’s guide, version 1.2, Dept. of Automatic Control and System Engineering, University of Sheffield, UK, 1994.
[10] D. Goldberg, “Genetic Algorithms in Search, Optimization and Machine Learning”, Addison-Wiley Publishing Company, Massachusetts, 1989.
[11] D. Gupta, and S. Ghafir, “An Overview of Methods Maintaining Diversity in Genetic Algorithms”, International Journal of Emerging Technology and Advanced Engineering, vol. 2, no. 5, pp. 56-60, 2012.
[12] K. Jones, “Comparison of Genetic Algorithms and Particle Swarm Optimization for Fermentation Feed Profile Determination”, Proc. CompSysTech’2006, Veliko Tarnovo, Bulgaria, June 15-16, pp. IIIB.8-1–IIIB.8-7, 2006.
[13] T. Pencheva, O. Roeva, and I. Hristozov, “Functional State Approach to Fermentation Processes Modelling”, Prof. Marin Drinov Academic Publishing House, Sofia, 2006.
[14] K. Schuegerl, and K.-H. Bellgardt (Eds.), “Bioreaction Engineering, Modeling and Control”, Springer-Verlag, Berlin Heidelberg New York, 2000.
IJSER © 2013 http://www.ijser.org