International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013 2114
ISSN 2229-5518
Phenotypic Investigation for Virulence factors of Pyocine producing Pseudomonas aeruginosa Isolated from Burn Wounds, Iraq. Huda H. Mohammad
Abstract: A total of 72 Transport swabs were collected from burned victims, who admitted to Burn unite in Al-Hilla General Teaching Hospital through 3/2013 to 5/2013. All Transport swabs were cultured on cetrimide agar and subjected for standard bacteriological procedures for bacteriological diagnosis.The results of P. aeruginosa isolation from burn victims revealed that 16/72 (22.22 %) were positive for pyocine producing P. aeruginosa culture. All pyocine producing P. aeruginosa were investigated for phenotypic detection of biofilm formation and virulence factors. The result show that (68.75%) of isolates were biofilm former (when investigated by TCP assay), also (68.75%) of isolates were positive for alkaline protease while all isolates had lipase and (87.5%) of isolates had phospholipase C, (81.25%) were positive for gelatinase. The results of antibiotics susceptibility test illustrate that, all isolates were sensitive for ciprofloxacin, aztreonam and polymyxin. Only one isolate was resist tobramycin and gentamycin and two and three isolates were resist to amikacin and imipenem respectively while all isolates were resist piperacillin.The current study concludes the high percentage of P. aeruginosa isolation, even though new techniques and solution for sterilization and disinfection, and had abundant capability to biofilm formation, virulence enzyme production and antibiotics resistance.
Keywords: Pseudomonas aeruginosa, Biofilm, Virulence factors, Antibiotics
1 Introduction:
—————————— ——————————
aeruginosa to adapt and thrive in wide variety of
environments is due in part to its extensive genetic
versatility, which contributes significantly to its potential
Pseudomonas aeruginosa is a non sporulating, gram
negative, oxidase positive motile bacterium with a polar
flagellum. It is ubiquitous microorganism widely
distributed in soil, water and on living hosts [1], and it can
thrive in hot baths, diluted sterilizers, disinfectants, contact lenses and catheters. P. aeruginosa is able to infect a wide variety of organisms including mammals, fish, invertebrates, and lower-order eukaryotes. It is a leading gram-negative pathogen that causes nosocomial infection which has received most attention [2]. P.aeruginosa has capacity to adapt easily to change in the environment. It needs a minimal nutritional requirement to grow and rapidly develop resistance to antibiotics and produce arsenal of virulence [3]. The remarkable ability of P.
————————————————
Huda H. Mohammad is currently working as lecturer in the department of
microbiology, College of Medicine, Babylon University
pH: 07602008590, Email-hudashmm@gmail.com
pathogenicity[4]. It is a significant opportunistic pathogen associated with skin and soft tissue infections, nosocomial pneumonia and sepsis [5].
The burn wound can be regarded as a culture medium, and a vascularity of the burn wound prevents the action of the blood-borne immune system. Furthermore, the use of invasive procedures bypassing the remaining mechanical and biological barriers increases the risk of influx of microorganisms. In addition, prolonged hospital stay and translocation of bacteria from the gastrointestinal organs also contribute to the increased risk for contracting infections, complicated further by the increased length of stay at the hospital and the use of more central venous catheters in burn patients [6]. Most of Pseudomonal infections are both invasive and toxinogenic. The ultimate Pseudomonas infection may be seen as composed of three distinct stages: (1) bacterial attachment and colonization; (2) local invasion; (3) disseminated systemic disease. However, the disease process may stop at any stage. Thepathogenicity of P. aeruginosa is multifactorial depends on numerous
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013 2115
ISSN 2229-5518
virulence factors including cell-associated factors and secreted factors[7].
The first step in P. aeruginosa infections is the colonization of the alerted epithelium, Adherence of P. aeruginosa to the epithelium is mediated by pili and flagella. Evidence also implicates LPS as an adhesive factor. P. aeruginosa produced viscous exopolysaccharaied (alginate) in upper airway of patient with cystic fibrosis. After colonization P.aeruginosa produce several extracellular virulence factors pyocin, alkaline protease, elastase, protease IV, heat-labile and heat- stable hemolysine, neuraminidase and exotoxins A,S,U,Y,T responsible for extensive tissue damage, blood stream invasion and dissemination, Many of these extracellular virulence factors are controlled by cell to cell signaling system [8]. Interaction between virulence factors and the host immune response determinate the severity and the type of infections. Adding to the problems of high incidence and infection severity, the resistance of P.aeruginosa to conventional antimicrobial treatment has increased over the past decade [9].It is important to stress that P. aeruginosa infections are difficult to treat because of the bacteria's intrinsic resistance to many antibiotics, owing to its low outer membrane permeability, and its ability to acquire new resistance mechanisms during antibiotic treatment [10].
Once colonization and infection being established, P.
aeruginosa becomes one of the worst pathogens of humans.
It is highly versatile in virulence and it is known to possess intrinsic multi-drug resistance capabilities. P.aeruginosa is the second most common bacteria cause nosocomial infections, accounting for 21% of cases. Incidences reported that 16% of nosocomial pneumonia, 12% of urinary tract infections, 17-26% of wound infections and 10% of septicemia are due to P. aeruginosa [10].
Intrinsic and acquired resistance makes P. aeruginosaas one of the most difficult organisms to treat and eradicate. P. aeruginosa represents an unusual phenomenon of antimicrobial resistance among prokaryotes, since practically all known mechanisms of resistance be found in this organism including decreased outer membrane permeability, pincillin binding protein modification, increased expression of efflux pumps system, alginate and enzymetic inactivation of antibiotics [10].
Although its intrinsically sensitive to ß-lactams (e.g., ceftazidime and imipenem, aminoglycosides (e.g., amikacin and tobramycin), and fluoroquinolones (e.g., ciprofloxacin and ofloxacin), resistance to thesetibiotics has emerged [11]. The outer membrane of P. aeruginosa is responsible for the
high resistance to many antimicrobial agents in comparison with other organisms in which there are some differences in
(LPS) composition and in that cation content of the outer membrane [12]. This study was conducted to investigate the virulence of the pyocine producing P. aeruginosa isolated from burned victims.
2 Materials and Methods:
2-1 Specimens Collection and Bacterial identification
The transport swabs were collected from burned victims before washing procedure and cultured on cetrimide agar and subjected for standard bacteriological procedures for bacteriological diagnosis [13].
3-Phenotypic detection of virulence factors:
3-1 Alkaline Protease Detection:
A single colony of an overnight growth from Nutrient agar were cultured on skim milk agar by picking of the colony, incubated at 37 C° for 24- 48 hr. The appearance of cleared hydrolysis zone indicates of positive test, as described by [14]
3-2 Lipase Detection:
Tween 20 agar was inoculated with a single colony of an overnight growth from nutrient agar incubated for (1-5) days at 37°C. Turbid zone around colonies with change to blue color after addition of the CuSO4 .5H2O reagent, indicates a positive result [15].
3-3 Phospholipase (Lecithinase) Detection:
The specific media for detection phospholipase was inoculated with a single colony of overnight culture from nutrient agar, incubated for (24-72hr) at 37C°. Changing the color of precipitation zone around the colonies from white to brown color considered a positive result [15].
3-4 Gelatinase Detection:
The specific media for detection gelatin liquefaction is inoculated with a single colony of overnight culture from nutrient agar and incubated for 24 hr at 37 C° of incubation period by placing the tube in refrigerator for (30)min. A positive result was indicates if the media was not solidify which refer to ability of bacteria to produced gelatinase [15].
4- Biofilm formation Assay:
The biofilm formation assay was achieved according to the Tissue culture plate method (TCP) assay (also called semi quantitative microtiter plate test (biofilm assay) described by [16]. The results were interpretated depending on the classification of bacterial adherence and biofilm formation by TCP method [17].
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013 2116
ISSN 2229-5518
5- Antimicrobial susceptibility test:
This test was performed on Muller-Hinton agar with the antibiotics discs according to [18]. Antibiotics inhibition zones were measured. Zone sizes were compared to standard to determine the susceptibility or resistance of organism to each antibiotic according to [18] criteria. Results:
The results of P. aeruginosa isolation from burn victims revealed that 16/72 (22.22 %) were positive for pyocine producing P. aeruginosa figure (1). This result was in harmony with those gathered from another studies in Iraq as in [19], found that (31.57%) were positive for P. aeruginosa. [20] in Mosul city, found that P. aeruginosa account for (5.7%) of clinical and environmental samples.
The results revealed that (68.75%) of isolates were biofilm former(when investigated by TCP assay. These results were in agreement with [21] who reported that (87.5%) of P. aeruginosa isolates had the ability to form alginate biofilm. [22] found that (66%) of P. aeruginosa isolates produce alginate biofilm. Bacteria in natural habitats usually grow as biofilms, organized communities of cells embedded in an extracellular polysaccharide matrix and attached to a surface.
phospholipids and has been shown to participate in the pathogenesis of P. aeruginosa. Hemolytic phospholipase
can suppress the host neutrophil oxidative burst response [9].
As regards the results of antibiotics susceptibility test illustrated in the fig. 4 show that, all isolates were sensitive for ciprofloxacin, aztreonam and polymyxin. Only one isolate was resist tobramycin and gentamycin and two and three isolates were resist to amikacin and imipenem respectively while all isolates were resist piperacillin.
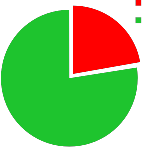
positive culture P. aeruginosa negative culture P. aeruginosa
22.2%
77.8%
Fig. 1. Percentage of P. aeruginosa isolation among burn victims.
The results of the phenotypic detection of virulence
factors of P.aeruginosa display that it has many virulence factors associated with its pathogenicity. Concerning the results of virulence factors of the P. aeruginosa isolates, fig. 3 show that (68.75%) of isolates were positive for alkaline protease while all isolates had lipase and (87.5%) of isolates had phospholipase C, (81.25%) were positive for gelatinase.
In relation to alkaline protease production the results
outlined that (68.75%) of isolates had alkaline protease and this results like those of [22] who found that (85%) of the P. aeruginosa isolates had alkaline protease. [23] stated that the vast majority of P.aeruginosa strains were shown to possess alkaline protease.
Regarding the results of lipase production the results show that all isolates were positive for lipase. This result
100.00%
50.00%
0.00%
68.75%
biofilm former
31.25%
non biofilm former
was higher than those documented by [24] who found that, only (42.85%) of isolates were positive for lipase.
The results of the current study were similar to those of the [25], [26] who reported that most P. aeruginosa isolates had phospholipase C. Phospholipase C more specifically hemolytic phospholipase C targets eukaryotic membrane
Figure. 2. Percentage of Biofilm formation among P. aeruginosa
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013 2117
ISSN 2229-5518
Gelatinase Phospholipase C Lipase
Alkaline protease


81.25%
87.50%
100.00%
68.75%
In addition, misuse of antibiotics and relaxation in general hygienic measures are associated with increasing infections
with these. The universal nature such as the ability to survive in the moist environment and resistance to many antibiotics make P. aeruginosa a common pathogen in intensive care units of the hospitals [28] ,[ 29].
The TCP method was found to be most sensitive, accurate
and reproducible screening method for detection of biofilm formation by P. aeruginosa and has the advantage of being a quantitative model to study the adherence of this bacterium on biomedical devices [17]. Crystal violet is a basic dye known to bind to negatively charged molecules on the cell surface as well as nucleic acids and polysaccharides, and therefore gives an overall measure of the whole biofilm. It has been used as a standard technique for rapidly accessing cell attachment and biofilm formation in a range of Gram positive and Gram-negativebacteria [30] , [ 31].
In the biofilm, the bacteria are resistant to the effects of antibiotics and phagocytosis from neutrophils. As planktonic (free living, non-biofilm) cells typically express many more immunogenic traits, these cells are vulnerable

Figure. 3. Virulence factors of P.aeruginosa isolates.

100%
100%
50%
to host defenses. Those bacteria remaining in the biofilm
continue to propagate and are metabolically active. This biofilm colony serves as a reservoir that may never be eradicated if the normal host defenses are not restored [32]. Within the biofilm, the bacteria are protected against the numerous surrounding PMNs, and they exhibit a remarkable tolerance to antibiotic treatments [33] , [34].
12.50%
0% 0% 6.25%
0%
18.75%
0% 6.25%
Alkaline protease interferes with fibrin formation
implicated in fibrin lysis, and inactivates important host defense proteins such as antibodies, complement and
CIP PI PB TOB AK AT IMP CN
Fig. 4. Antibiotics resistance of P. aeruginosa isolates
Discussion:
The percentage of P. aeruginosa is flexible and may be accredited to drug overuse, to the hospital policy in management of such cases. Furthermore, geographic climatic and hygienic factors may also be associated with the relative variability of results among different area. The high prevalence of P. aeruginosa in community may be related to the increasing numbers of the immunocompromised patients in our population due to different diseases and contaminations of the environment of the country [27].
cytokines [35]. The importance of this enzyme is the
increasing the production level of some virulence factors such as toxin A and elastase in which this enzyme gives a primary material for growth and spread of bacteria [36].
In vitro studies of the function of cells mediating the
immune response revealed that lipase significantly
inhibited monocyte chemotaxis and chemiluminescence. Furthermore, a combination of lipase and PLC led to a dramatic increase in the formation of 12- hydroxyeicosatetraenoic acid (12-HETE) from human platelets [37]. Thus, the present state of knowledge classifies the P. aeruginosa lipase as an important virulence factor which induces its harmful effects in combination with other bacterial enzymes, in particular PLC. These pathogenicity factors may activate or suppress the immune response from various cells and thus contribute to the pathophysiology of P. aeruginosa infection in a distinct manner [38]. Aminoglycoside-resistance in Pseudomonas spp. is primarily due to the genetic expression of enzymes responsible for the
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013 2118
ISSN 2229-5518
modification of the aminoglycosides. There are three specific classes of aminoglycoside-modifying enzyme that have been identified, the N’acetyltransferases, phosphotransferases and adenyltrnsferases [39].
Resistance to imipenam in P. aeruginosa is significantly associated with reduced uptake of the agent. These results from the loss or reduced expression of the OprDporin and also is associated reduced susceptibility to meropenem. Resistance to imipenem may also arise via Amp Chyperproduction and/or overexpression of the intrinsic efflux systems [40].
The outer membrane of the P. aeruginosa is intrinsicall
poorly permeable to many classes of compound with a permeability coefficient range from 10 to 500 fold lower than that of E. coli. P. aeruginosa OmpH has been demonstrated to be an antibiotic resistance related protein. The role of OprF in antibiotic resistance remains controversial. It has been suggested that of loss of this protein may be involved in the multiple antibiotic resistance phenotype and it has been proposed that OprF has a role in antibiotic uptake [41].
Conclusion:
The current study concludes that, the high percentage of P. aeruginosa isolation, even though new techniques and solution for sterilization and disinfection, and had abundant capability to biofilm formation, virulence enzyme production and antibiotics resistance.
References:
[1] B.O. Akanji, J.O. Ajele, A. Onasanya and O. Oyelakin, "Genetic Fingerprinting of Pseudomonas aeruginosa Involved in Nosocomial Infection as Revealed by RAPD-PCR Markers" Biotech. Vol. 10, no. 1, pp.70-77,
2011.
[2] S. Meenakumari, S. Verma, A. Absar, and A. Chaudhary, "Antimicrobial susceptibility pattern of clinical isolates of P.aeruginosa in an Indian cardiac Hospital", Internat. J. Engineer. Sci.Tech., vol. 3, no. 9, pp. 7117-7124,
2011.
[3] T. Strateva, B. Markova, Ivanova, and I. Mitov, "Distribution of the type III effector proteins-encoding genes among nosocomial Pseudomonas aeruginosa isolates from
Bulgaria", Ann. Microbiol., vol. 60, pp.503-509, 2010.
[4] S. Finnan, F.J. Morrissey, P. O'Gara, and E.F. Boyd, "Genome Diversity of Pseudomonas aeruginosa Isolates from Cystic Fibrosis Patients and the Hospital Environment". J.Clin.Microbiol., vol. 42, no. 12, pp. 5783-5792, 2004.
[5] F.H. Damron, and J.B. Goldberg, "Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa", Mol.Microbiol., vol. 84, no.4, pp.595–607, 2012.
[6] K.Tang, L. Jian, Z. Qin, L. Zhenjiang, M. Gomez, and M.
Beveridge, "Characteristics of burn patients at a major burn center in Shanghai", Burns, vol. 32, pp.1037–1080,
2006.
[7] O. Karatuna, and A. Yagci, , "Analysis of quorum sensing – dependent virulence factor production and its relation with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates", Clin. Microbiol. Infect. Dis., vol.16, pp.1770-1775, 2010.
[8] A. Cotar, M. Chifiriuc, S. Dinu , M. Bucur, C. Iordache, O.
Banu, O. Dracea, C. Larion, and V. Lazar , "Screening of Molecular Virulence Markers in Staphylococcus aureus and Pseudomonas aeruginosa Strains Isolated from Clinical Infections", Int. J. Mol. Sci., vol.11, pp.
5273-5291, 2010.
[9] E. Kipnis, T. Sawa, and J. Wiener-Kronish, "Targeting mechanisms of Pseudomonas aeruginosa pathogenesis", Médecineet maladies infectieuses, vol. 36, pp. 78-91,
2006.
[10] T. Strateva, and D. Yordanov, "Pseudomonas aeruginosa - a phenomenon of bacterial resistance", J. Med. Microbiol., vol.58, no.9, pp.1133-1148, 2009.
[11] J. Sekiguchi, T. Asagi, T. Miyoshi-Akiyama, A. Kasai, Y.
Mizuguchi, M. Araake, T. Fujino, H. Kikuchi, S. Sasaki, H. Watari, T. Kojima, H. Miki, K. Kanemitsu, H. Kunishima, Y. Kikuchi, M. Kaku, H. Yoshikura, T. Kuratsuji, and T. Kirikae, "Outbreaks of Multidrug- Resistant Pseudomonas aeruginosa in Community Hospitals in Japan", J. Clin. Microbiol., vol.45, no. 3, pp.979-989, 2007.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013 2119
ISSN 2229-5518
[12] G. MacDonnell, and D. Russell, "Antiseptics and Disinfectants: Activity, Action and Resistance", Clin. Microbiol. Rev., vol.12, no.1, pp.147-179, 1999.
[13] J.F. MacFaddin, "Biochemical tests for the identification of medical bacteria:, 3rd ed., The Williams and Wilkins- Baltimor, USA. 2000.
[14] H.J. Benson, "Microbiological Applications: Laboratory Manual in General Microbiology", (8th ed). Complete version. McGraw-Hill. U.S.A. 2002.
[15] J.G Collee, A.G. Fraser, B.P. Marmino, and A. Simons, "Mackin and McCartney Practical Medical Microbiology". 14thed. The Churchill Livingstone, Inc. USA. 1996.
[16] G.D. Christensen, W.A. Simpson, J.A. Younger, L.M.
Baddour, F.F. Barrett, and D.M. Melton, "Adherence of cogulase negative Staphylococi to plastic tissue cultures: a quantitative model for the adherence of staphylococci to medical devices". J. Clin. Microbiol.,vol. 22, pp.996–
1006, 1985.
[17] T .Mathur , S. Singhal, S. Khan, D.J. Upadhyay, T. Fatma, and A. Rattan, "Detection of biofilm formation among the clinical isolates of staphylococci: An evaluation of three different screening methods". Ind. J. Med. Microbiol.,vol. 24, no.1, pp. 25-29, 2006.
[18] Clinical and Laboratory Standards Institute (CLSI) "Performance standards for antimicrobial-susceptibility testing. Informational supplement. M 100-S22., Wayne, Pannsylvannia, vol. 32, no.3, pp.1-184, 2012.
[19] H.H.S. Al-Mamori, "Characterization of Pseudomonas aeruginosa Isolated from Patients and Hospital Environment in Hilla City. M.Sc. thesis. thesis in Microbiology. College of Medicine. Babylon Univ.
2011.
[20] K.A. Al-Mashhadani, "Study on Diagnosis and Pathogenesis of Pseudomonas aeruginosa Isolated from Different Sources in Mosul City". Ph D thesis. Thesis in Microbiology. College of Science. Mosul Univ., 2004.
[21] S.H. Moteeb, "Quantitative and Qualitative assays of
bacterial biofilm produced by Pseudomonas aeruginosa
and Klebsiella spp". J. Al-Anbar univ. pure. Sci., vol.2, no. 3, 2008
[22] R.H. Saleh, "Immunological and molecular Study on Pseudomonas aeruginosa isolated from clinical samples in Babylon Province", Babylon University, Medicine faculty. Iraq, Ph.D. thesis. 2012.
[23] R.S. Bradbury, L.F Roddam, A. Merritt, D. W. Reid , and A.C. Champion, "Virulence gene distribution in clinical, nosocomial and environmental isolates of Pseudomonas aeruginosa", J. Med. Microbiol., vol. 59, pp. 881-890,
2010.
[24] B. Zouaoui, A. Bouziane, and R.G Bachir, "Production, optimization and purification of lipase from Pseudomonas aeruginosa. Afr. J. Microbiol. Res., vol.6, no.20, pp. 4417-4423, 2012.
[25] K. Wolska, And P. Szweda, "Genetic Features of Clinical Pseudomonas aeruginosa Strains. Polish .J.Microbiol., vol.58, no. 3, pp.255-260, 2009.
[26] I. Mitov, T. Strateva, and B. Markova "Prevalence of virulence genes among Bulgarian nosocomial and cystic fibrosis isolates of P.aeruginosa". Brazilian
.J.Microbiol., vol.41, pp. 588-595, 2010.
[27] H. Memmel, A. Kpwal-Vern, and B. Latenser, "Infections in
Diabetic Burn Patients. Dia. Care.,vol. 27, pp. 229-233,
2004.
[28] P. Mahar, A.A. Padiglione, H. Cleland, M. Eldho, W.H.
Paul, and Wasiak, "Pseudomonas aeruginosa bacteraemia in burns patients: Risk factors and outcomes. Burns, vol. 36, no.8, pp. 1228-1233, 2010.
[29] Sh. Taherzadeh, F. Soheili, Z. Deilami, H. Salimizand, A.
Heidari, S. Beiranvand, and E. Kalantar, "Incidence of nosocomial infections caused by P.aeruginosa among burn patients at Kurdistan province .Glo.Res .J. Microbiol.vol. 2, pp. 035-0.38, 2011.
[30] D. Djordjevic, M. Wiedmann, and LA. McLandsborough, "Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol., vol. 68, pp. 2950-2958, 2002.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013 2120
ISSN 2229-5518
[31] C. Matz, D. McDougald, A.M. Moreno, P.Y. Yung, F.H.
Yildiz, and S. Kjelleberg, "Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholera", Proc. Natl. Acad. Sci. USA., vol. 102, pp.16819-16824, 2005.
[32] G.W. Lau, D.J Hassett, B.E Britigan, "Modulation of lung epithelial functions by Pseudomonas aeruginosa.Trends Microbiol., vol, 13, pp. 389–97, 2005.
[33] R.M. Donlan, and J.W. Costerton, "Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev., vol. 15, pp. 167-193, 2002.
[34] E. Drenkard, "Antimicrobial resistance of Pseudomonas aeruginosa biofilms". Microbes Infect., vol. 5, pp. 1213–
1219, 2003.
[35] L. Smith, B. Rose, P. Tingpej, H. Zhu, T. Conibear, J.
Manos, P. Bye, M. Elkins, M. Willcox, S. Bell, C. Wainwright, and C. Harbour, "Protease IV production in Pseudomonas aeruginosa from the lungs of adults with cystic fibrosis", J. Med. Microbiol., vol. 55, pp.
1641-1644, 2006.
[36] M.A.H. Jasim, "Genetic Study of Biofilms and Free Pseudomonasspp. Causing Urinary Tract Infections. M. Sc.thesis. College of Science. Babylon Univ., 2010.
[37] B. KÖnig, K.E. Jaeger, and W. Ko¨nig, "Induction of inflammatory mediator release (12- hydroxyeicosatetraenoic acid) from human platelets by Pseudomonas aeruginosa". Int. Arch. Allergy Immunol.,vol. 104, pp. 33–41, 1994.
[38] P. Friedl, B. Ko¨nig, and W. Ko¨nig, "Effects of mucoid and non-mucoid Pseudomonas aeruginosa isolates from cystic fibrosis patients on inflammatory mediator release from human polymorphonuclear granulocytes and rat mast cells", Immunology, vol. 76, pp. 86–94, 1992.
[39] E. B. Hirsch, V. H. Tam, C.A. Rogers, K. Chang, J.S.
Weston, J. Caeiro, and K.W. Garey, "Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes" Expert. Rev. Pharmacoeconomics
Outcomes Res., vol.10, no.4, pp. 441-451, 2010.
[40] S. Kouda, M. Ohara1, M. Onodera, Y. Fujiue, M. Sasaki, T.
Kohara, S. Kashiyama, S. Hayashida, T. Harino, T.Tsuji, H. Itaha, N. Gotoh, A. Matsubara, T. Usui, and M. Sugai, "Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima". Med.J. Antimicrob.Chemother., vol.64, no.1, pp. 46-51, 2009.
[41] A. Dötsch, T. Becker, C. Pommerenke, Z. Magnowska, L.
Jänsch, and S. Häussler, "Genomewide Identification of Genetic Determinants of Antimicrobial Drug Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother., vol.53, no.6, pp. 2522-2531, 2009.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 7, July-2013
ISSN 2229-5518
2121
I£ER lb) 2013
http://www.ijserora



