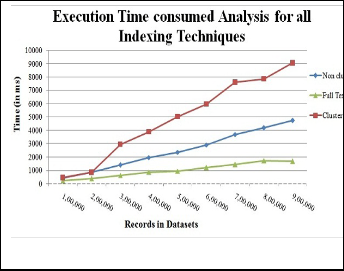
indexing is less than full text index but better than cluster index.
Fig. 2. Indexes Execution time analysis
International Journal of Scientific & Engineering Research, Volume 3, Issue 3, March-2012 1
ISSN 2229-5518
Abstract— Biomedical data warehouse is the central management system having pool of DNA/RNA protein datasets where protein identification, matching and searching are important operations. The traditional data warehouse was designed in such a manner that it can efficiently manage transactional data which is highly dominated by numerical information where as in biomedical data warehouse textual and non transactional information is encountered. The dataset which contains text data is accessed over the network on the daily basis and performance issue arises. The objective of this paper is to propose an indexing technique based on time complexity and index space complexity for data warehouse used in application of biomedical field. The performance evaluation of three data warehouse queries is focused in this paper by comparing techniques used mostly with FULL TEXT indexing technique and to observe the results of variable size dataset with respect to time and space complexity.
Index Terms— Cluster Index, Non-Cluster Index, Full Text Index, Bio Medical, Data Warehouse.
ata warehouse is the centralized storage of data which improves strategic decision making with coherent views of data because it provides the ability to make quick,
well-informed decisions which are critical to competitiveness and growth for an organization. Biomedical databases are dif- ferent from traditional data warehouse as it contains non transactional information like bimolecular (protein, RNA, DNA, lipid, carbohydrate), organelles, nomenclature
,reference resources and web directories which can size up to petabytes (1000 terabytes). The main operations on biomedical databases includes searching of protein and matching of cer- tain patterns of data which is very complex and tedious task to handle with because finding a particular pattern of RNA/DNA in warehouse can even take days, so pattern matching or searching becomes difficult in the systems as it takes considerable amount of time and memory consumption depending upon the queries which are complex and iterative [6]. Consider scenario of forensic science which deals with crime issues which are increasing day by day, a large amount of data is accumulated which is needed to be accessed in least amount of time when complex queries are executed in present biomedical data warehouses which is a major challenge. The ability to answer these complex queries efficiently depends upon a major factor 'Index'. A database index is a special re- presentation of information about objects that improve search- ing.
This means performance of these complex queries and ad hoc queries will be greatly enhanced if right index structures are built on columns of biomedical data warehouse which con- tains DNA protein sequence of characters having large length.
•Amit bansal is currently pursuing master’s degree program in Internet technol- ogy from Lovely Professional University, India, PH-09888602049.
E-mail: bansal.amit1989@gmail.com
•Sankalap Arora has completed his M.tech from Lovely professional University, PH-09779039097. E-mail: sankalap.arora@gmail.com
Indexing of a data warehouse is complex and if there are few indexes, the data loads quickly but the query response is slow.
If there are many indexes, the data loads slowly and there will be more storage requirements but the query response is good. This is true with large tables and complex queries that involve table joins. Considerable amount of time is taken by the query to be processed is more due to large size of both tables and attributes. Index’s space and time play an impor- tant role in choosing an indexing technique in data warehouse.
Usually if the space used by an index is large then the re- sults are achieved in short time and on the other side, if the space used by the index space is small then the results are achieved in greater amount of time. So there is a tradeoff be- tween the time consumed and the space used by a particular index.
Bioinformatics is the application of computer technology of management of biological information like DNA which con- tains the sequence of large sequence/chain of A, T, C, G cha- racters having length of 80 characters approx. In forensic science DNA is used to identify felons or in other cases find relatives of one person having its DNA which is very difficult task.
Searching of proteins from biomedical data warehouse and matching of protein sequence of the users with each other and then analysis of searched information becomes difficult be- cause of the increasing number of users which further increase size if DNA datasets[5].
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 3, March-2012 2
ISSN 2229-5518
1. Response time
2. Searching time/Scan time
3. Memory Usage
Searching problems efficiently and less response time can be
attained by choosing a good index on biomedical data ware-
house. There are number of existing indexing techniques
which have many advantages and drawbacks on each other.
Following are the factors which are used to determine which indexing technique should be used on data warehouse from various existing indexing techniques.
dinality of data which can be further classified into three cate-
gories:
a) High cardinality: It can be unique ID number of employees.
b) Normal cardinality: It can be combination of first and last
name of employee.
c) Low cardinality: It can be value in which male or female
employee is classified.
dered in data warehouse to identify indexing technique be-
cause some indexes are created in less amount of time but
some take considerable amount of time.
to generate the range and count range sizes, according to
which the index should be selected.
1. Index size should be less.
2. Index should support with indexes of another data ware-
house indexes.
3. Index should take less memory for processing.
4. Creation time of index should be least.
5. Indexes should be able to work with joins & ad-hoc queries.
There are existing techniques of indexing for searching the data that is clustered index, non clustered index and Full Text index .Brief description of these techniques with their advan- tages and disadvantages over each other is given below:-
It is a type of index in which the data is arranged in distinct order (in sequence) which means clustered index determines the physical order of data in table. It is beneficial when there is need to access the records sequentially or in the reverse order. There can only be one clustered index per table, because the data rows themselves can only be sorted in one order. There are row locators which is clustered index key on the row. The only time the data rows in a table are stored in sorted order is when the table contains a clustered index. If a table has no clustered index, its data rows are stored in a heap [1]. Advantages
1. Block reads are less because data is arranged in sequential order.
2. Data is accessed faster from tables
1. Inserts and updates take longer time with clustered index
2. Cluster index is avoided when there are concurrent inserts
on almost same clustering index value.
The data of records are present in random order but the logical ordering is created by index. The index partition is one but when there are more partitions, then every partition is kind of B-tree structure which contains the indexes. The phys- ical order of the rows is not the same as the index order [4]. These indexes are mostly created on column where queries like JOIN, WHERE, and ORDER BY clauses are used and bet- ter for those tables whose values are modified frequently. Advantages
1) Use of non clustered indexes results in retrieving data faster as it retrieve everything from index pages without bothering bookmark lookup into table to read data row.
2) Data in columns need not to be sorted.
3) Each table can have as many as 249 non clustered indexes
1) Each index takes up disk space and drag on data modifica-
tion resulting in serious issues regarding storage require-
ments.
2) By using non clustered index the number of rows return are
less.
3) When using non cluster index there is no saving from one
row to other as successive entries of non clustered index refer-
ence rows on disk pages that are likely to be far apart.
Full text indexing provides the feature of full text queries for character based data where there is LOB (large object) kind of columns is present like varchar which gives better efficiency because it gives accurate result due to division of characters strings into tokens and each token searching is fast like search- ing in documents. When there are fewer documents then full text searching scans tokens in documents called serial scan- ning. If the large document is present for searching keywords then full text has to follow two steps:
a) Indexing b) Searching
Indexing means scanning the text from documents often called index of the text searched and searching phase only search references rather than large documents which makes full text indexing more efficient than other indexing techniques. Advantages
1. It is less time consuming and returns results in seconds ra- ther than minutes.
2. It uses small set of T-SQL statements and functions. Exam- ple: CONTAINS and FREETEXT.
3. Full text indexes also support boolean modes like AND/OR
criteria.
1. Fails to identify repeated words which consume additional time in processing similar words.
2. Sequence of searching is not supported by full text index.
3. It gives the results of queries as large collection of data.
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 3, March-2012 3
ISSN 2229-5518
In this section techniques are implemented and tested on the basis of memory consumed with dataset of different sizes. These techniques are full text index, cluster index and non cluster index. The graphical representation shows all index- ing techniques with respect to different datasets on different type of queries.
The dataset which is used for analysis is taken from world- wide protein data bank [10].The dataset of 44,00,000 records is divided into different sub datasets to identify the variation in size and calculation of execution time of different indexes.
Creation time of various indexing techniques corresponding to different datasets has been calculated and on basis of these values, indexing techniques have been analyzed on factors like CPU time and creation time.
CPU time is the combination of compilation time and execu- tion time. The performance of indexing techniques is shown graphically. It is observed that the full text index is better than other indexes because it takes less CPU time in comparison to others. The cluster index’s consumption of CPU time in- creased gradually as the number of records in datasets is in- creased. The time taken by full text index is 2000 ms approx and cluster index took 9000 ms approx. It is observed that cluster index CPU time hike from 2000 ms to 5000 ms approx in range 2 to 3 lakh records. Non cluster index is in mid range and time is 5000 ms approx.
indexing is less than full text index but better than cluster index.
Fig. 2. Indexes Execution time analysis
Cluster index takes much time than other indexes and on the contrary, it takes less memory nearly 500 KB approx. Full text index consumes more memory and on the contrary, it gives best performance than others. After 5 lakh records in full text index, the memory size varies in less but after 8 lakh, memory size hike from 3000 KB to 5000 KB.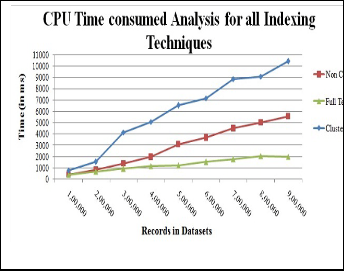
Fig. 3. Indexes Memory consumption analysis
Fig. 1. CPU time analysis of Indexing techniques.
Initially, the execution time for all indexing techniques is below to
1000 ms from 1 to 2 lakh record. Full text index execution time is below to 2000 ms but there is fast hike of time in the cluster index i.e
9000 ms approx till 9 Lakhs record. The performance of non cluster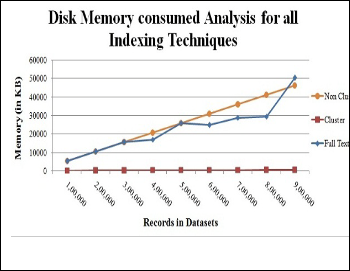
SQL stands for Structured Query Language which is used to communicate with a database using SQL commands such as "Select", "Insert", "Update", "Delete", "Create", and "Drop" [8]. Critical issue in data warehouse is that answer complex and iterative queries efficiently because most of the queries contain a lot of join operations and retrieve large number of records as results and needs several hours or days to process. Basically there are three basic SQL statements or queries which often
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 3, March-2012 4
ISSN 2229-5518
used in data warehouse:- a) Full table scan Query b) Exact Match Query
c) Statistical Type Query
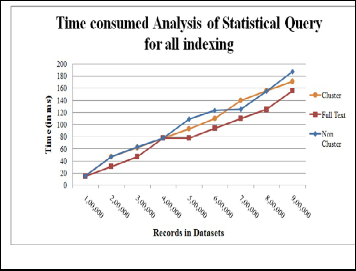
Count is the function to calculate the number of records in the column of table contained in dataset. Counting of records on full text index columns takes less time than other indexes. At 4 lakh , all in- dexes are at same stage i.e. 80 ms. The non cluster index had taken more time than the cluster index and in between, the time varies from high to low as shown in figure 4.
Cluster index gives better results in searching of exact match- ing of string and results good performance even the records in datasets are increasing.
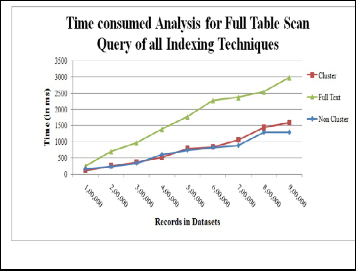
Full table scan means searching in every row of tables con- tained in datasets. SQL keyword LIKE is used for searching entire tables [9]. In this, non cluster is efficient rather than oth- er indexes because according to results, it takes nearly 1000 ms which is less than other indexes. Full text indexed columns consume much time, i.e. 3000 ms approx where as non cluster index is good enough for full table scanning.
Fig. 4. CPU time analysis for Statistical Query
Fig. 6. CPU time analysis Full Table Scan Query
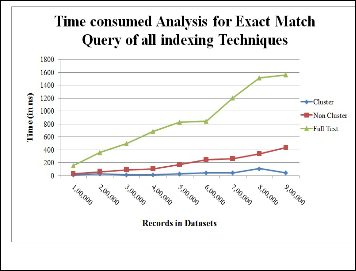
In SQL, ‘where’ key word is used for searching of exact keyword in dataset [7]. The full text time consumption is in- creasing rapidly but on the contrary, Non Cluster index time consumption is much less nearly 400 ms approx.
Fig. 5. CPU time analysis for Exact Match Query
Nodes contain the values, condition or can be another structure. When the nodes take decision on the basis of some input contains in nodes, then it becomes decision tree and use- ful for easily analysis of inputs, predict values and possible outcomes of values [6]. Decision tree mapped the output val- ues corresponding to input values. Decision tree is classified in different stages and can be used that approaches with differ- ent requirements [2].
1. Top-Down Approach
2. Bottom-Up Approach
3. Pruning Approach
4. Hybrid Approach
Decision tree works according to conditions and corres-
ponding, the best approach should select best results. It can be
used for artificial neural network, decision values, signal
processing, and statistical analysis where data can be explored
with the different scenarios [3] [6].
1. Generalization: Input and output is mapped taking con-
sideration independent input to dependent output.
2. Classification: The data contains the predefined belong-
ing class or not and benefit is that, specific operation can be
performed.
3. Description: Large volume of Information can be reduced
to compact form and it reduces the memory problems.
IJSER © 2012 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 3, Issue 3, March-2012 5
ISSN 2229-5518
In DNA Protein datasets, the feature detection can be classi- fied and corresponding, indexes of classes can build. This ap- proach will reduce the memory problems, CPU time, and ex- ecution time. This index scheme will enhance the performance of data warehouse.
In today's scenario biomedical data warehouse plays a cru- cial role in order to perform important operations like protein identification, matching but challenge is large size of biomedi- cal data warehouse as it contains RNA/DNA which encapsu- late string of large length. Different indexing techniques has been used and analyzed using different types of queries on different size of datasets in biomedical data warehouse in or- der to perform operation in efficient manner. Full text index- ing provides better performance than cluster and non cluster indexes but on the contrary, it consumed much memory com- paratively other techniques which is costly for large data warehouse. It can be improve by create new data structure of index which contains splitting criteria of characters and it must be memory and cost effective with timely information.
[1] Sankalap Arora, Priyanka Anand, Kirandeep Singh,” An Efficient
Indexing Technique Used In Telemedicine Data Warehouse”, (2010)
[2] Safavian, S.R., Landgrebe (1991), “D.A survey of decision tree classifier methodology”, Systems, Man and Cybernetics, IEEE Transactions, Page 660-674.
[3] Sreerama K. Murthy (1998),” Automatic Construction of Decision Trees from Data: A Multi-Disciplinary Survey”, Association for Computing Machinery
[4] Shawana Jamil and Rashda Ibrahim (2009) “Performance analysis of
Indexing Techniques in Data Warehousing”, Emerging Technologies,
2009. ICET 2009 International Conference, Page 57-61
[5] S.Thabasu Kannan, Dr.K.Iyakutti (2009) “A Clustered Indexing Method for Optimizing the Query for Biological Databases”, GCC Conference & Exhibition, 2009 5th IEEE, page 1-6.
[6] Khalid Jaber, Rosni Abdullah and Nur’Aini Abdul Rashid (2009) “Indexing Protein Sequence/Structure Databases Using Decision Tree: A Preliminary Study”, Information Technology (ITSim), 2010
International Symposium, IEEE Computer Society Conference,
[7] Vikram vaswania (2004) “The Complete Reference, MySQL”, Tata
McGraw-hill Publishers Ltd., New Delhi.
[8] Julie C. Meloni (2007), “Sams Teach Yourself PHP, MySQL and Apache All in One, Third Edition”, Sams Publisher., United States of America.
[9] Chris Leiter, Dan Wood, Paul Turley (2007), “Beginning SQL Server™ 2005 Administration”, Wiley Publishing, Inc., Indianapolis, Indiana.
[10] http://www.pdb.org
IJSER © 2012 http://www.ijser.org