International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1276
ISSN 2229-5518
Novel Anticancer enzyme from Marine
Bacteria to fight against neoplastic
cancer-Acute lymphoblastic leukemia†
Pallavi Rudrapati*
![]()
Abstract Recently there has been an explosion of information about novel bioactive compounds that have been isolated from marine microbes in an effort to further explore the relatively untapped marine microbes and their secondary metabolites for drug discovery. New therapeutic agents are urgently needed to treat medicinal needs that are currently unmet .The biodiversity of marine microbes and the versatility of their bioactive metabolites has not been fully explored. L-asparaginase therapy, alone is finding increased success in the management of acute lymphocytic leukemia’s. Large amounts of purified enzyme and increasing recognition of successful therapy will generate increased demand. The production of L-asparaginase by using bacteria has attracted great attention owing to their cost effective and eco-friendly nature. In search of efficient microorganisms potential for production of anticancer enzymes, bacteria isolated from mangrove soil of Nizampatnam Guntur district were screened for L-asparaginase .The screened isolates were biochemically characterized and identified as pseudomonas species. In the present study one of the potential Pseudomonas strain was selected and identified as Pseudomonas aeruginosa AVP17 by 16s rRNA partial sequence. Effect of pH, temperature, carbon source, salt tolerance was studied for growth optimization and maximum production of enzyme.
KEYWORDS Mangrove soil, Marine bacteria, L-Asparaginase, neoplastic cancer, Acute lymphoblastic leukemia, Pseudomonas aeruginosa, 16S rRNA partial sequence.
![]()
1. INTRODUCTION
Cancer is defined as uncontrolled
division of cells. Acute lymphoblastic leukemia is cancer of white blood corpuscles (WBC) characterized by the excessive multiplication of malignant and immature WBC (lymphoblast) in bone marrow. Treatment of acute leukemia includes chemotherapy, steroids, radiation therapy and intensive combined treatments including bone marrow or stem cell transplants.![]()
• Pallavi Rudrapati
Research scholar
Department of Microbiology Acharya Nagarjuna University Guntur,A.P,India pallavirudrapati3@gmail.com Ph.+919573412117,+918008294049
Among these, chemotherapy is most preferred.
The drugs being employed for treatment
includes prednisolone, dexamethasone,
vincristine, asparaginase, daunorubicin, cyclophosphamide, cytarabine, etoposide, thioguanine, mercaptopurine, hydrocortisone, methotrexate etc[1]. Although, variety of drugs is available today, their efficacy in treatment of cancers at third and fourth stage is doubtful. The side effects caused by these chemotherapeutic agents are many such as infertility, secondary neoplasm, nausea and vomiting, immunosuppression etc. The term “acute” means that the leukemia can progress quickly, and if not treated, would probably be fatal within a few months. Lymphocytic means it develops from early (immature) forms of lymphocytes, a type of white blood cell. L-
asparaginase is the first enzyme with anti-
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1277
ISSN 2229-5518
leukemic activity to be intensively studied in
human beings [2]. It is an enzyme drug of choice used in combination therapy for treating acute lymphoblastic leukemia in children [3,4,5,6,]. Since 1922, L-asparaginase has been considered as a therapeutic agent against malignant tumors [7,8,9]. It was further shown that growth of normal cells was not dependent on L- asparagine. L-asparaginase was introduced in the therapeutics due to the fact that in a significant number of patients with acute leukemia, particularly lymphocytic, the malignant cells are dependent on an exogenous source of L-asparagine for survival. Normal cells, however, are able to synthesize L- asparagine and thus are less affected by its rapid depletion produced by treatment with the enzyme L-asparaginase.Amino acid degrading enzymes are important chemotherapeutic agents for the cure of some types of cancers.
The manufacture or processing of enzymes for
use as drug is an important facet of today’s
pharmaceutical industry[10](Cassileth, B., 1998). The microbes are the best sources of L- asparaginase because they can be cultured easily and the extraction and purification step is also convenient, facilitating the large scale production [11] The common side effect of this medication is [12].
Recent times, L-asparaginase emerged as potent
health care agent for the treatment of acute lymphocytic leukemia [13,14,15,16] because tumor cells cannot synthesize L –asparagine and hence take L-asparagine from blood circulation or body fluid. More over L-asparaginase is biodegradable, non-toxic and can be administered at the local site quite easily. Prior to the discovery of the anti-leukemic and antitumor effects of the enzyme L-asparaginase, no qualitative difference in nutritional requirement between cancer and normal cells
were known and there were no
chemotherapeutic agents which were qualitatively specific for any type of cancer cell.
All chemicals used in this study investigation were of analytical grade and procured from Sigma
(USA), Hi – media (India) and Merck (India).
Soil samples were collected from mangrove soil sediment of Nizampatnam of Guntur district at a depth of 3 ft. and placed in zip locked plastic bags at 40 C .The soil contained 3.8% of organic matter and pH8.8.
on agar plates of modified M-9 medium [17].
1 gm. of soil was separately suspended in 9 ml of physiological saline soil in a flask and placed on an orbital shaker (at 100 rpm) at room temperature (28± 20C) for 1 hr. At the end of shaking the soil samples were serially diluted up to 10-6 with physiological saline. 104-106 dilutions were placed on modified nutrient agar medium containing Fluconazole (antifungal antibiotic) by pour plate technique and incubated at 280C .The most prominent colonies were isolated maintained on Nam slants at 40c for further studies.
The medium contained Na2HPO4.2H2O, 6.0 g; KH2PO4, 3.0 g; NaCl, 0.5 g; L-Asparagine, 10.0 g; 1mol- MgSO4.7H2O, 2.0 ml; 0.1 M solution of CaCl2.2H2O, 1.0 ml; 20% glucose stock, 10.0 ml;
agar 20.0 g. per liter of distilled water. The
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1278
ISSN 2229-5518
medium was supplemented with 0.005% phenol
red dye (prepared in ethanol) and the pH was adjusted to 6.2 using 1N HCl. Plates were then incubated at 37°C for 24-48 hrs. A set of tubes was also run as a control without L-asparagine. The strains having potential for L-asparaginase production were selected on the basis of pink zone formation and retained for further screening.
Morphological characters such as shape and color of the colonies were examined. Grams staining and motility were also done. Isolates were biochemically analyzed for the activities of oxidase, catalase, MR-VP test, starch hydrolysis and gelatin hydrolysis, indole production, hydrogen sulphide test, nitrate reduction, sugar fermentation and citrate utilization. The results were compared with Bergey’s Manual of Systematic Bacteriology. Out of 46 bacterial isolates 9 selected bacterial isolates showing excellent L-asparaginase activity were characterized on the basis of morphological and biochemical characteristics and the results were interpreted according to Bergey's Manual of Determinative Bacteriology & PIBWIN software version 19.2[18,19].
Pure culture of AVP 17 bacterial isolate was grown until log phase achieved and genomic DNA was isolated essentially[20].The amplification of 16S rRNA gene was done by using universal bacterial primer 1492R (5´- TACGGYTACCTTGTTACGACTT-3´) and 27F (5´ AGAGTTTGATCMTGGCTC AG- 3´) as per the conditions[21].The PCR product was sequenced at Macrogen South Korea. The sequences obtained were compared with those from the GenBank using the BLAST
program[22]and Phylogenetic trees
reconstructions were obtained by the Neighbor
joining method 1000 bootstrap replicates were performed to assess the statistical support for each branch in the tree[22,23].
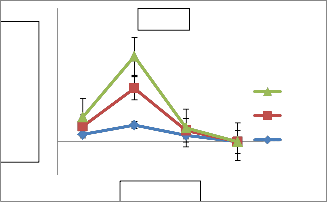
Effect of different temperature ranges (25⁰c,37⁰c,50⁰c,100⁰c), different incubation periods(24 hrs,48 hrs,72 hrs), pH values(7,9,10,12,14), salinity concentrations(0.5%,5%,10%,15%,20%), different carbon sources(21), different Nitrogen sources(11), different amino acids(16) and metal ions/mineral salts (51) on enzyme production was studied.
The rate of hydrolysis of L-Asparagine was determined by measuring the ammonia released using Nessler’s reaction[24].The color reaction was allowed to develop for 10 min and the absorbance read at 480 nm with a spectrophotometer. The ammonia liberated was extrapolated from a curve derived with ammonium sulphate. One unit (U) of L- Asparaginase was defined as that amount of enzyme which liberates 1 μ mole of ammonia per minute under the assay conditions [25].
The culture filtrate was filtered through whatmann No. 1 filter paper and centrifuged at
8000 rpm for 10 min at 4⁰C. The culture filtrate
(crude enzyme) was brought to 45 per cent saturation with ammonium sulphate at pH 8.4 and kept overnight in a cold room at 4⁰C. It was thereafter subjected to centrifugation at 8000 rpm for 10 min at 4⁰C. The precipitate was discarded, while the supernatant was brought to
70 per cent saturation with ammonium sulphate and centrifuged at 8000 rpm at 4⁰C for 10 min. The precipitate from this step was collected and stored at 4⁰C.
To measure the kinetics of L-asparaginase, Michaelis constant (Km) and Maximal velocity
(Vmax) of the partial purified enzyme was
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1279
ISSN 2229-5518
determined. They are one of the important
parameters for the evaluation of the potential usefulness of the enzyme for anti-leukemic therapy. They were determined using L- asparaginase as substrate in the range of 0.01M-
1M concentration. Each reported velocity I the
mean of at least three measurements. The apparent Km was determined [26].
Bacterial strains were isolated from soil samples collected from Nizampatnam of Guntur district. L-Asparaginase bacterial strains were identified by pink colored colony on agar medium with phenol red as an indicator. whereas one plate maintained as control as uninoculated (Figure 1) and another plate was inoculated with selected bacterial strain AVP 17 (Figure 2).Microbial strain AVP 17(from Nizampatnam soil sample), was selected and further characterized by morphological, physiological and biochemical studies. The isolated strain, AVP 17 was Gram- ve and rod shaped colony (Table 1) colonies with fluorescent blue colour under UV light (Figure 3). According to the data of Bergy’s manual of systemic bacteriology,[27] based on morphological physiological and biochemical characteristics(Table 1) the bacterial strain AVP
17 was classified to be Pseudomonas sp. A 1466 bp PCR product of gene was amplified from the genomic DNA of AVP 17 .A sequence similarity showed that the 16srDNA gene sequence of AVP 17 had 99% similarity to the 16srDNA of Pseudomonas aureginosa strain and Pseudomonas species PPB2(AC:HM771657).The sequence was blast in NCBI and for analysis. Based on phylogenetic analysis revealed that AVP 17 was closely related to Pseudomonas aureginosa (Figure
4) and sequence was deposited in NCBI as Pseudomonas aureginosa AVP 17 with accession number KF527831. Optimization of physico chemical parameters like pH, temperature and salinity plays an important role in production of
L-asparaginase.
Optimization revealed that AVP 17 showed
maximum production (1.10 IU/ml) at 37⁰c after
48 hrs incubation (Fig.5). Production of L- asparaginase varies with incubation period at
37⁰c indicating optimization of incubating period also exhibit a significant role(Fig.5) Enzyme production of AVP 17 showed variation at different pH. L-asparaginase production was found to be maximum (16.19 IU/ml) at pH 9 and observed to be gradually decreased beyond pH
12(Fig. 6). Percentage of NaCl concentration also
effect the L-asparaginase production of AVP
17.The production was inversely related with increasing concentration of NaCl and found to be maximum(19.11 IU/ml) at 0.5% concentration(Fig.7).Earlier literature revealed that L-asparaginase production in Streptomonas albidoflavus was observed to be high(11.9 IU/ml) at 40⁰c at pH 7.5 Isolate AVP 17 showed maximum l-asparaginase production with Dextrose (166.25 IU/ml) and Urea (89.1 IU/ml), the carbon and nitrogen source respectively.(Fig.8)Amino acid Glutamine and sodium tungstate were observed to be potential inducers for L-asparaginase production of 66.07
IU/ml and 774.91IU/ml respectively(Table 2).
After optimization Dextrose, Urea, Glutamine and sodium tungstate were selected as potential inducers and enhancers. An attempt was made to evaluate the extent of improvement in production of l-asparaginase in the modified formulated production media with necessary inducers and enhancers. 5 folds of enhancement in L-asparaginase production observed with modified production medium indicating highly significant improvement so far observed. Enzyme kinetics, Vmax and Km values were studied at different substrate concentrations (0.01-1 M) and at different incubation periods (24 hrs, 48 hrs and 72 hrs). At 24 hrs of incubation, Vmax value of L-asparaginase is
133.82 and Km value is 0.5. At 48 hrs , Vmax is
193 and Km value is 0.2 and at 72 hrs AVP 17
showed 27.2 Vmax value and 0.03 Km value.
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1280
ISSN 2229-5518
Enzyme kinetics study revealed that L-
Asparaginase of AVP 17 showed High Vmax at
0.7 M substrate. (Fig.11). Enzymatic activity of the strain AVP17 measured at physiological temperature showed lower Km values. The Km values obtained were closer those of some mesophilic L-asparaginase of earlier studies.
Production of L-asparaginase using different microbial systems has attracted much attention, owing to the cost-effective and ecofriendly nature. A wide range of microorganisms including bacteria, fungi and yeast have proved to be the beneficial sources of this enzyme. L- asparaginase is an important natural product that possesses a broad spectrum of antitumor activity. It has been successfully applied to the treatment of several diseases such as lymphocyte sarcoma and leukemia. It has been
proved that L-asparaginase from Escherichia coli
and Erwinia carotovora has anti-neoplastic activity against cancer and is being used as anticancer drug. But, it is observed that the action of enzyme is coupled with some side effects. Moreover, the yield of enzyme was not enough to fulfill the demand of the drug. Solid state fermentation is being adopted all over the world for the production of the enzyme as it has many advantages over submerged fermentation. In this concern the isolated bacterial strain AVP17 was potent and produces high yield of L-asparaginase under optimized conditions.So, it created the need to discover new sources and techniques to enhance the yield and decrease the side effects of the enzyme. Enzyme isolated from different sources has different optimized conditions for production and activity. Recombinant work and formulation of enzyme is also in progress, yet there is still a long way to go.
Figure 1 | Figure 2 |
|
|
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1281
ISSN 2229-5518
AVP 17 showing Fluorescence
under UV

Isolated strain | AVP 17 |
Morphological analysis | |
Gram staining | Negative |
Shape | Short Rod |
Physiological analysis | |
Starch hydrolysis | Positive |
Lipase | Negative |
Urease | Positive |
H2S Production | Positive |
Protease | Positive |
Gelatin hydrolysis | Negative |
HCN Production | Positive |
Litmus | Positive |
Biochemical analysis | |
Indole | Negative |
Methyl red | Negative |
Vogues Proskeur | Negative |
Citrate | Positive |
Catalase | Positive |
Oxidase | Positive |
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1282
ISSN 2229-5518

4
3
2
1
0
25˚c 37˚c 50˚c 100˚c
-1
72hrs
48hrs
24hrs
Figure 6: Influence of pH on enzyme activity
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1283
ISSN 2229-5518
30 AVP 17
20
10
0
-10
7 9 10
pH
12 14
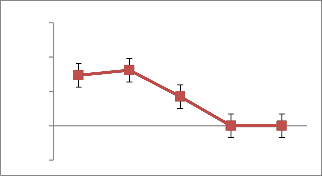
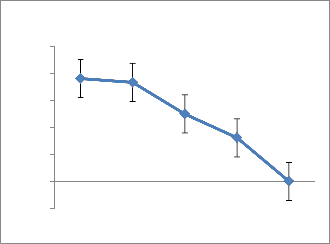
Figure 7: Influence of NaCl on enzyme activity
25
20
15
10
5
0
0.50% 5% 10% 15% 20%
-5
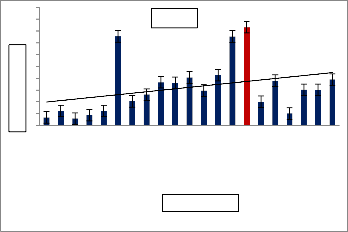
Figure 8: Influence of carbon sources on enzyme activity
200
180
160
140
120
100
80
60
40
20
0
AVP 17
Carbon sources
Figure 9: Influence of various Nitrogen
sources on enzyme activity
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1284
ISSN 2229-5518
100
80
60
40
20
0
AVP 17
Nitrogen sources
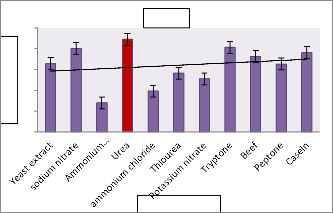
Table 2 : Influence of various Metals/Mineral
Sources on enzyme activity
S.N0 | Mineral salts | IU |
1 | calcium oxalate | 50.31 |
2 | ammonium ceric sulphate | 48.3 |
3 | ammonium sulphate | 69.33 |
4 | cupric sulphate | 34.82 |
5 | ammonium carbonate | 54.41 |
6 | ammonium oxalate | 63.08 |
7 | ammonium borate | 74.46 |
8 | Ammonium Chloride | 39.28 |
9 | Calcium chloride | 45.04 |
10 | potassium chloride | 174.1 |
11 | ammonium thiocyanate | 74.46 |
12 | cobaltous acetate | 74.01 |
13 | Manganese chloride | 82.41 |
14 | alluminium sulphate | 81.83 |
15 | alluminium nitrate | 84.5 |
16 | cupric nitrate trihydrate | 84.64 |
17 | barium bromide | 66.42 |
18 | lead acetate | 79.68 |
19 | copper-phosphate | 82.67 |
20 | Sodium chloride | 69.59 |
21 | Mercuric chloride | 71.33 |
22 | Lithium chloride | 72.58 |
23 | Magnesium sulphate | 81.56 |
24 | Creatinine | 74.91 |
25 | Manganese sulphate | 57.54 |
26 | copper sulphate | 50.13 |
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1285
ISSN 2229-5518
27 | ferric citrate | 76.2 |
28 | Zinc | 76.33 |
29 | Silver nitrate | 76.25 |
30 | Selenium | 84.06 |
31 | Barium chloride | 75.08 |
32 | Potassium ferricyanide | 65.8 |
33 | Ferric choride | 72.32 |
34 | Ferrous sulphate | 84.59 |
35 | Potassium thiocyanate | 52.5 |
36 | Calcium carbonate | 84.15 |
37 | Sodium thiosulphate | 53.52 |
38 | Sodium tungstate | 774.91 |
39 | ammonium molybdate | 79.86 |
40 | Potassium acetate | 52.94 |
41 | ammonium ferrous sulphate | 53.08 |
42 | Lithium sulphate | 63.48 |
43 | Potassium nitrate | 50.84 |
44 | potassium carbonate | 54.86 |
45 | sodium hydroxide | 40.13 |
46 | sodium acetate | 44.1 |
47 | zinc sulphate | 65.49 |
48 | Sodium hydrogen carbonate | 58.43 |
49 | Zinc chloride | 57.23 |
50 | Trisodium citrate | 60.13 |
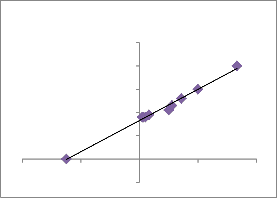
Figure 11: Line weaver Burk Plot
Line weaver Burk Plot
0.1
0.08
y = 0.0013x + 0.0332
R² = 0.9908
0.06
0.04
0.02
0
-40 -20 0 20 40
-0.02
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1286
ISSN 2229-5518
Authors are grateful to UNIVERSITY GRANTS COMMISSION–SPECIAL ASSISTANCE PROPGRAMME-BASIC SCIENCE RESEARCH New Delhi India for sanctioning Junior Research Fellowship to Pallavi Rudrapati, Department of Microbiology Acharya Nagarjuna University Guntur Andhra Pradesh INDIA.
The authors declare that there is no conflict of interests regarding the publication of this paper.
,Vol. 5(1), Jan, 2012.
2. SAVITRI, ASTHANA N and AZMI W, Microbial L-Asparaginase: A potent antitumor enzyme. Indian Journal of Biotechnology. 2:184-194, 2003.
Successful treatment of acute lymphoblastic leukaemia with L- asparaginase induced intracranial hemorrhage to activated recombinant factor VIIa in a child. Pediatr. Hematol. Oncol, 23, 339-345, 2006.
promising chemotherapeutic agent, Crit. Rev. Biotechnol, 27, 45-62, 2007.
541, 1961.
highly selected standard risk childhood acute lymphoblastic leukaemia. Haematologica, 90, 186-1191, 2005.
Statistical and evolutionary optimisation of operating conditions for enhanced production of fungal Lasparaginase, Chemical Papers, 65 (6)
798–804, 2011.
12. SARQUIS M,I, Production of Lasparaginase by filamentous fungi: Mem. Inst. Oswaldo Cruz, 99: 489-492,
2004.
13. BESSOUMY EIAA and SARHAN M, Production, isolation and purification of L-asparaginase from Pseudomonas aeruginosa 50071 using solid state fermentation. J Biochem Mol Biol .2001;
37: 387-393.
28, 465-474, 2007.
15. GUPTA R, A rapid plate assay for screening of L-asparaginase producing microorganisms, Letters in Applied Microbiology,24: 23-26, 1997.
PAREKH, P.P, Isolation, optimization
and production of L-asparaginase from
IJSER © 2014 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 5, Issue 12, December-2014 1287
ISSN 2229-5518
coliform bacteria. Asian J.
Biotechnol.2010; 2: 169-177.
17. GULATI, R AND SAXENA, R.K, A rapid plate assay for screening of L- asparaginase producing microorganisms. Letters in Applied Microbiology, 24: 23-26, 1997.
manual of determinative bacteriology,
9th ed. Williams and Wilkins Co., Baltimore,USA, I BN:13-978 0 63- 063-2,
1994.
1995.
2005; 89(1), 177-180.
1990.
Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 24:1596-1599,
2007.
24. MASHBURN L T and WRISTON J C, Tumor inhibitory effect of L- asparaginase from E. coli, Arch Biochem Biophys , 105,450-452, 1964
13-18, 1964.
anti-leukemic enzyme L-asparaginase from marine actinomycetes by solid state and submerged fermentation, Purification and Characterization,Trop.J.pharm,
8(4),353-360, 2009.
R.PALLAVI and G.NAGA RATNASUPRIYA, Characterization of L-Asparaginase Producing Bacteria from Mangrove Soil International Journal of Chem Tech Research, Vol.5, No.1, 109-112, 2013.
IJSER © 2014 http://www.ijser.org