Inte rnatio nal Jo urnal o f Sc ie ntific & Eng inee ring Re se arc h, Vo lume 3, Issue 3, Marc h-2012 1
ISS N 2229-5518
Multiple gene dysfunctions lead to high
cancer-susceptibility: evidences from a whole-exome sequencing study
Soniya priyadharishni.A.K, Dr.M.Sridhar, Dr.M.Rajani
24,545 SNPs w ere detected. 10,868 (44.27%) SNPs w ere w ithin coding regions, and 1,077 (4.38%) located in the UTRs. 3367 genes w ere hit by
4480 non-synonymous mutations in CDS w ith truncation of 30 proteins; and 10 mutations occurred at the splice sites that w ould generate diff erent protein isof orms. Substitutions or premature terminations occurred in 132 proteins encoded by cancer -associated genes. CARD8 w as completely loss; ANAPC1 w as pre-translationally terminated f rom the transcripts of one allele. On the Ras -MAPK pathw ay, 18 genes w ere homozygously mutated. 15 grow th f actors/cytokines and their receptors, 9 transcription f actors, 6 proteins on WNT signaling p athw ay, and 16 cell surf ace and extracellular proteins may be dysf unctioned. Exome sequencing made it possible f or individualized cancer therapy.
—————————— ——————————
t has always been bothering physicians to choose correct drugs as the anticancer effects are completely different among patients. This is caused by not only the multiple ge- netic mutations in human cancers but also a wide variety of single nucleotide polymorphisms (SNPs) of individuals. Mu- tations in exons, such as mutations on H-RAS, p53 and APC genes, are often found to cause human cancers. Up to date, 73 genes with germline mutations and 412 genes with germline or somatic alterations, including amplification, deletion, rea r- rangement and point mutations, have been shown to be i n- volved in human cancers in the Cancer Gene Census of Cancer Genome Project database (CGC/CGP). In the Atlas of Genetics and Cytogenetics Oncology and Haematology (AGCOH) da- tabase, there are 766 annotated genes that are genetically ass o- ciated with cancers and other 3,000 other genes are functiona l- ly involved in the process of cancer development. Although a great advance has been achieved for early diagnosis of human cancers and anticancer drug development, the mobility of ca n- cer cases is increasing while the average mortality almost re- mains consistent in the last decades. The random use of anti- cancer drugs largely neutralized the attempts of anti cancer treatment; and cancer is still the second killer of human dis- eases. Therefore, it is urgently needed to develop genome-
based individualized cancer therapy and care.
It is well known that the whole exome constitute only about
1% of the human genome but harbor the major of mutations
contribute to cancer development. Therefore, combined with
bioinformatics analysis, targeted exome sequencing technol o- gy would be a good and practical strategy to largely reduce the cost and labor load. It would also have a great potential to expand our knowledge of rare mutations in cancer develop- ment and to accelerate the functional studies of cancer- associated genes. Using high susceptibility of cancers as proof-
of-concept, we observed that 132 genes, which have been shown to be important for cancer development, dysfunctioned or functionally alternated. Of them, only 11 genes were ger- mline-mutated according to CGC/CGP database; while the mutations of other 121 genes were newly identified in ger- mline in cancer patient.
A very unique cancer patient, a chinese women (YH2), was recruited in this study. She underwent breast cancer, gallbladder adenocacinoma and lung cancer at 41, 63 and 66, respectively. She died of recurrence of gallbladder adenocac i- noma in liver at 68. The tumors were removed by surgery at the diagnosis and tumor types were determined by histoch e- misty assays after surgery. There was no family history of can- cers. Informed comment was obtained from the patient for this study, and the study was approved by the ethic committee of The Chinese University of Hong Kong.
The strategy for exome sequencing was similar as described by Ng et al. In brief, shotgun libraries were generated from 10 ug of blood leukocytes purified genomic DNA (gDNA) using the standard Illumina protocols. The fragments of size 150 -200 bp were isolated after electrophoresis on 6% PAGE and hybri- dized with NimbleGen 2.1M-probe sequence capture array, in which oligos were fixed to cover the human exomes (RefSeq, NCBI 36.3, 33.92 Mb). The captured exomes were applied for direct single-end sequencing on an Illumina Genome Analyzer II. The average read for each probe is 75 bases. Sequences were then aligned to the reference (RefSeq, hg18, 19 and YH1) using SOAP aligner, and the mapped bases, depth, coverage and the base distribution were analyzed.
IJSER © 2012
Inte rnatio nal Jo urnal o f Sc ie ntific & Eng inee ring Re se arc h Vo lume 3, Issue 3 , Marc h-2012 2
ISSN 2229-5518
SNPs were called by SOAPsnp based on the alignments with HapMap database. For each site within the exome targeted region, only copy number <1.5 of the surrounding area was allowed and the depth should range from 10X to 200X. Finally, a Q20 threshold was used to filter unreliable SNPs. After ex- cluding known substitutions from the potential mutations available, the SNPs were annotated and the genes involved in cancer development were revealed by comparison of our data with CGC/CGP and the AGCOH database.
For the single reads we produced, the short in -dels <4 bp were also identified by S0APaligner2 in a gap tolerable mode. Local alignments were performed with our custom perl scripts.
MAGEE2 and IL17RB) have been recorded to have genetic associations with cancer; while 11 other cancer-associated pro- teins, for the first time, were observed to be mutated in the germline. Particularly, MAGEE2, which has been shown ge- netic association in melanoma and hepato-cellular carcinoma, was truncated at N-terminal by homozygous mutations. CARD8, a key factor for the recruitment of caspase in apopto- sis pathway, was almost completely loss in the patient. ANAPC1, a key components of ana -phase promoting complex that play crucial roles in cell mitosis and protection of the i n- tegration of chromosomes from separa tion, truncated >70% by a heterozygous mutation at Gln465. Some important proteins on the RAS-MAPK signaling pathway, including G protein coupled receptor 1 (GRP1), tyrosine kinase (MAP2K3), and protein tyrosine phosphatase (PTPN11), also prematurely terminated.
![]()
TABLE 1: NONSENSE MUTATIONS (ST, ST OP; JMML, JUVENILE MYE- LOMONOCYT IC LEUKEMIA; AML, ACUTE MYELOGENOUS LEUKEMIA; MDS, MYELODYSPLAST IC SYNDROME)
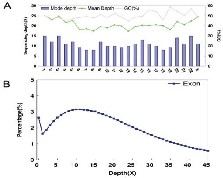
Our sequencing strategy was similar to the one published by
Ng et al recently but with a larger coverage (33.92 instead of
26.6 megabases). The average sequencing depth was 21.1 (Figure 1). The total reads were about 1.97 Gagabases (GBs) which covered 97.36% of the reference. With SOAPaligner
![]()
Name T Y P e
Muta- tion
Position
(stop)
f ull length (aa)
Function G enetic association with di s- ease(s)
software, 87.92% of bases were aligned to the reference (build
131,10/03/26, hg18 and hg19) and YH genome sequence. The
Functional associated with cance rs
mismatch rate was 0.65%, indicating the data was in high s e- | ANAPC1 | H | CAG>TAG | Q465 | 1926 | anaphase |
quencing quality. We detected total 24,545 SNPs. Among | E | promoting | ||||
them, 10,874 (44.3%) SNPs located in the coding regions and T complex | ||||||
142 (0.6%) SNPs located in the UTRs. There were 23,604 SNPs were shared among YH1 and dbSNPs, while 941 SNPs were | G PR1 | H | CGA>TGA | R236 | 355 | signal trans- |
142 (0.6%) SNPs located in the UTRs. There were 23,604 SNPs were shared among YH1 and dbSNPs, while 941 SNPs were | E | duction |
newly identified in the patient after comparative analysis of
SNPs in the captured exome. Among them, 8091 SNPs
(42.81%) were homozygous. 3058 genes were hit by 4480 non- synonymous mutations in the coding sequences (CDS). 10 mutations displayed at spice sites, and 8 small in/dels were identified.
Fig: 1 Targeted capture exome sequencing. A. Chromosome depth and GC distribution in targeted capture exome regions. X axis stands f or each chromosome, Y1 axis presents the sequencing depth and YH2 axis is the
GC proportion in exon capture region of each chromosome.
We detected 33 nonsense mutations that caused truncation of
30 proteins (Table 1). We found only 3 proteins (PTPN11,
![]()
T ASCC3 H E![]()
T MAP2K3 H E
T PT PN11 H E![]()
T MAG EE2 H O
M CARD8 H O![]()
M
AB CA10 H E T
![]()
IL17RB H E T
CAG>TAG Q87 111 signal trans- duction
CAG>TAG Q73 318 tyrosine kinase, signal transduction
TAT> TAG Y197 593 protein tyro-
sine phosph a- tases
GAG>TAG E120 523 signal trans-
duction
TGT> TGA C10 432 caspase recruitment
CGA>TGA R1322 1544 drug transport
CAG>TAG Q484 502 cytokine receptor
JMML, AML, MDS melano ma, HCC
rheumatoid arthritis
intestinal inflamma- tion
IJSER © 2012
Inte rnatio nal Jo urnal o f Sc ie ntific & Eng inee ring Re se arc h Vo lume 3, Issue 3 , Marc h-2012 3
![]()
ISSN 2229-5518
![]()
UB E2NL H E T
FTHL17 H
E T![]()
TP53RK H E T
Others
![]()
SPATA21 H E T
PZP H
E
TTA> TGA L89 153 ubiquitin ligation
GAG>TAG E148 183 ferritin heavy polypeptide- like protein
CGA>TGA R152 254 TP 53-
regulating kinase
CGA>TGA R467 470 spermatog e- nesis
CAA>TAA Q598 1483 proteinase inhibitor
Missensense mutations hit over 3,000 proteins. After aligned with the CGC/CGP and AGCOH databases, we observed i m- portant substitutions (most likely causing function alterations) occurred in 132 proteins, which strongly associated with can- cer development (Table 3). Among them, 45 have been record- ed as somatic mutations and only 11 recorded as germline mutations in cancer patients in the CGC/CGP database. Tota l- ly 121 cancer-associated genes were newly found to display mutations in germline; some mutations would cause signifi- cant function alterations.
![]()
TABLE 2: HOMOZYGOUS MUTAT ION(S) IN GENES ST RONGLY (EIT HER GENET ICALLY OR FUNCT IONALLY) ASSOCIAT ED WIT H CARCINOGENE- SIS (*, HET EROZYGOUS MUTAT ION)
T UNC5CL H E
T
TCTE1 H
CAG>TAA Q12 519 NF-kB inhibi- tor
CAG>TAG Q460 502 t-complex-![]()
Name FL (aa)
RAS-MAP K
signaling pathway
mutations Name FL (aa) Mutations
Wnt signaling pathway
E associated - T testis -
expressed 1![]()
EML4 981 K283E AP C 2843 V1822D ENPP 2 865 S493P CD97 786 R318Q
EP HA1 976 M900V DKK2 259 R146Q
ASCC3 H
E
CAG>TAG Q87 2203 RNA helicase![]()
FNIP 1 1166 G76C, Q648R
DKK3 350 R335G![]()
T
ZN F75D H
CGA>TGA R331 511 transcriptional
GP R103 431 L344S Growth factors/cytokines and their recep-
tors/signal transducers
E T
DK FZp54 H
7 O M
factor
TGG>TGA W141 150 unknown
GP R112 3080 T1213N.S1
540P , F1791L, I276M*,
P 368H*
FGFR4 802 V10I, P 136L
LOC1496 H
![]()
43 E T
CGA>TGA R37 98 unknown![]()
GP R116 1346 T604M IGF2R 2491 R1619G, N2020
GP R142 462 H132N IL23R 629 Q3H, L310P
GP RC6A 926 P 91S MST1R 1400 Q523R(E), S1195G,![]()
MS4A12 H E T
CAA>TAA Q71 267 me mbr ane protein
GRP 115 695 K541N PP ARGC
1A![]()
R1335G(E)
798 G482S
OR2T5 H
E
CGA>TGA R24 315 olfactory
receptor
GRP 56 693 S281R TNC 2201 V295M*, Q539R,
V605I, E2008Q*
T
PZP H
CAA>TAA Q598 1483 pregnancy -![]()
KLK4 251 S22A*, H197Q
TNFRSF
10A
468 H141R, R209T, R441K
E zone protein![]()
T
KLK5 293 N153D TNFRSF
17
184 N81S
SLC6A18 H
TAC> TAG Y319 628 unknown![]()
KLK10 276 S50A, L149P *
TRAF3 568 M129T
KLK11 250 G17E P LEK2 354 S217C![]()
NIN 2046 Q1125P , G1320E
Cell cycle control![]()
ZN F75 H E T
ZN F80 H
E T
CGA>TGA R331 510 zinc finger protein
TAT> TAG Y245 273 zinc finger protein![]()
![]()
Apoptosis/anti -apoptosis Others CARD8 432 C10ST ASXL1 1541 L815P BCL2L2 194 Q133R CDH11 796 T255M*,![]()
![]()
M275I*,S373A
OP TN 577 M98K*, BRIP 1 1249 S919P
IJSER © 2012
Inte rnatio nal Jo urnal o f Sc ie ntific & Eng inee ring Re se arc h Vo lume 3, Issue 3 , Marc h-2012 4
![]()
![]()
ISSN 2229-5518
DNA repair/RNA
synthesis
K322E
COL1A1 1464 T1075A
y tion line p
e
![]()
ERCC5 1186 G1053R, G1080R, D1104H
FANCA 1454 T266A,A41
2V*,G501S
,P 643A*,G![]()
809D, T1328A*
DDX43 648 K625E,
Q629R
GOLGA5 731 A67G*,P 350L
LCP 1 627 K533E
LIFR 1098 D578N
ACSL3 H
e t

ADAM12 H o m
ADAM8 H o m
ADAMST5 H
719 L641H prostic cancer
1593 G48R
823 W35R, F657L
929 R614H,L
ATM 3056 N1983S MA- GEE2
523 E120ST(GAG>TA G)
e 692P
t
BUB1B 1049 R349Q MEN1 615 T546A![]()
![]()
Transcription factors NUT 1132 P 22L
ADAMTS4 H
o
1226 S538N
AFF3 1226 S538N P DE4DI P
2346 R25L*, A167T*, R681H*, C708R,
R1504Q*
m
AKAP 12 H
e
324 K118Q,K
1218I
multiple cancers, anti -angiogenesis![]()
CDX2 313 P 293S P MS2 862 P 470S*, T485K*, GATA2 480 A146T P 0U6F2 691 P 191L
Homozygous mutations displayed in 58 genes that may con- tribute to high susceptibility of cancers in this patient. Homo- zygous missense mutations occurred in 18 genes on RAS - MARK pathway, including G-protein coupled receptors (GPRs), tyrosine kinases and phosphatases (Table 2). On this pathway, heterozygous mutations hit 9 other genes, including AKAP12, CBLB, MAP2K3, MAP3K7IP1, PTPN11, PTPN21, TCL1B and USP6 (Table 3). Although the proteins encoded by these genes play critical roles in cells response to extracellular signalings; however, only EML4 and NIN were recorded s o- matic mutations in tumors in the CGC/CGP database. The second largest group (10 genes), which were hit by h omozyg- ous mutations, were growth factors/cytokines and their recep- tors. Although only mutation of TNFRSF17 was shown in the intestinal T-cell lymphoma in the database, the products of these genes are important to control cell growth and immune responses to infection and other human diseases including carcinogenesis. On the Wnt signaling pathway, besides APC, homozygous mutations of CD97, DKK2 and DKK3 most likely cause significant alteration of protein functions. The genetic alterations in tumors have not yet recorded. Apart from DDX43, the other homozy-gously mutated genes (ATM, BUB1B, ERCC5 and FANCA) for cell cycle control and DNA/RNA process were shown genetic association with ca- cinogenesis (Table 2). Besides function association, the ger- mline mutations of transcription factors (AFF3 and POU6F2) have not yet recorded. All 3 apoptotic/anti-apoptotic genes (CARD8, BCL2L2 and OPTN) were newly observed genetic alterations in cancer patients. This would enhance the somatic cells escaping from apoptosis during carcinogenesis.
![]()
G ene T FL (aa)1 Muta Somatic G erm

t
AKR1C4 H o m
ALOX12 H o m
ANAP C1 H
e t
AP C H o m
ASNS H
e t
ASXL1 H o m
ATF6 A
T F
6
ATM H o m
BCAS1 H
o m
BCL2A1 H![]()
e t
BCL9 H
e
324 S145C*, Q250R, L311V*
662 N322S
1926 Q465ST( GAC-
>TAC)
2843 V1288D colorectal, pan- creatic, desmoid, hepatoblastoma, glioma, other CNS cancers
561 V210E
1541 L815P MDS, CM ML
670 A145P , P 157S
3056 N1983S T-P LL
584 Q24K, V163A *
174 C19Y, N39K, G82D
1426 A218V B-ALL, Hodgkin
lymphoma
the same cancers as somatic mutations
leukemia, lymphoma, medullo b- lastoma, gliom
co- lon/breast/o![]()
IJSER © 2012
Inte rnatio nal Jo urnal o f Sc ie ntific & Eng inee ring Re se arc h Vo lume 3, Issue 3 , Marc h-2012 5

![]()
ISSN 2229-5518
t vary cancer, AML, leukemia, rhabdo- myosarca-
ma
DKK3 H o m
EML4 H
o
349 R335G gas- tric/lung/breast/p rostate/ovary cancer, glioma
980 K283E NSCLC
BMP R1A H
e t
BRIP 1 H
o m
531 P 2T breast cancer AML, leukemia, breast cancer
1249 S919P
m
ENPP 2 H o m
EP HA1 H
o
865 S493E
976 V160A
BUB1B H o m
1049 R349Q colorectal cancer, breast cancer
gastrointes- tinal neo p- lasia,
rhabdo-
myosarco-
m
ERCC2 H
e t
759 K751N skin basal cell, mel a- noma,
SKC,
CABC1 H
e t
CARD8 H o m
CARS H
e
ma
647 H85Q
432 C10st
(TGT-
>TGA)
879 A774T ALCL
ERCC5 H o m
FGFR2 H
e t
FGFR4 H
o m
1186 G1053R, G1080R, D1104H
820 M186T gastric, endome-
trial cancer, NSCLC
802 V10I
skin basal cell, SKC, melano ma
t
CBLB H
e t
CCND3 H
e t
CD97 H
o m
Het N466D AML
292 S259A MM
785 R318Q
FLT3 H
e t
FNIP 1 H o m
FTHL17 H
e t
FXYD5 H
992 T227M, D358V
1165 G76C, Q648R
183 E148st
(GAG -
>TAG)
178 S35A,
AML, ALL
CDH11 H o m
795 T255M*, M275I*, S373A
aneurismal bone cycs
o m
GATA2 H
R176H *
479 A146T AML
CDX2 H o m
CENP F H o m
COL1A1 H o m
COL1A2 H o m
DDX43 H o m
DKK2 H
o m
313 P 293S AML
3113 R2729Q, R2943G, N3106K
1465 T1075A
1365 P 549A dermato fibrosar- coma protub e- rans
647 K625E
259 R146Q
o m
GGH H
e t
GOLGA5 H o m
GP R1 H
e t
GP R103 H o m
GP R112 H
o m
317 C6R
730 A67G*,P
350L
355 R236st
(CGA -
>TGA)
431 L344S
3080 276M*,
P 368H *,T
1213N, S1540P ,
papillary thyroid
IJSER © 2012
Inte rnatio nal Jo urnal o f Sc ie ntific & Eng inee ring Re se arc h Vo lume 3, Issue 3 , Marc h-2012 6
ISSN 2229-5518
GP R116 H o m
GP R142 H o m
GP RC6A H o m
GRP 115 H o m
GRP 56 H o m
HTATIP 2 H o m
IGF2R H o m
JAG2 H
e t
KLK10 H o m
KLK4 H o m
KLK5 H o m
LCP 1 H o m
LIFR H o m
LOX H
o
F1791L![]()

1345 T604M
462 H132N
925 P 91S
694 K541N
692 S281R
276 S231R
628 Q3H, L310P
1237 E501K
275 S50A, L149P *
250 S22A*, H179Q
292 N153D
626 K553E NHL
1097 D578N salivary aden o- ma
417 R158Q
MGC34647 H
e t
MMP 10 H
e t
MMP 11 H o m
MMP 17 H
o m
MMP 20 H o m
MMP 26 H
o m
MMP 27 H
o m
MMP 8 H
o m
MMP 9 H
e t
MST1 H
e t
MST1R H
o m
MTHFR H
e t
MYEOV H
e t
266 Y213st
(TAC-
>TAG)
475 D81Y
486 A38V
602 A182T
482 K18T*, V275A,T
281N
260 K43E
512 M30V
467 K87E
706 Q279R
724 R108Q, R122Q
1399 Q523R/E, S1195G, R1135G/ E
655 A222V
312 V159A, R198Q,
G271R
breast cancer
m
LOXL2 H o m
LOXL4 H o m
MAP 2K3 H
e
773 M570L
755 R154Q
317 Q73st
(CAG-
MYH11 H
e t
MYST3 H
e t
NBN H
e t
1937 N1899S AML
2003 L134S
753 E185Q
t
MAP 3K7IP1 H
e t
MEN1 H![]()
o m
>TAG)
503 C235W
614 T546A P ara thyroid tumors
NIN H o m
NOTCH2NL H
e t
2045 Q1125P , G1320E
235 S67P ,![]()
P 133L, T158I, S181R,
MP D
ma rginal zone lymphoma, DLBCL
IJSER © 2012
Inte rnatio nal Jo urnal o f Sc ie ntific & Eng inee ring Re se arc h Vo lume 3, Issue 3 , Marc h-2012 7
![]()
ISSN 2229-5518
NQO1 H
e t
NSD1 H
e t
NUP214 H
e t
NUT H o m
OP TN H o m
P 2RX7 H o m
P BX1 H
e t
P 188H
239 Q139W
2695 S726P AML
2090 P 754S AML
1131 P 22L lethal midline carcinoma
576 M98K *, K322E
594 Y155H *, R270H *, E496A *, N568I
429 G21S P re B-ALL
t![]()
REL H
e t
RHOD H o m
RHOT2 H
e t
ROS1 H
e t
SDC1 H o m
SELE H
e t
SERP INB5 H
e
618 N424S many cancers and other di s- ease
210 C134R
617 A88T, R245Q
2347 T145P
310 L136Q
371 S303R
374 S176P , I319V
P DE4DIP H o m
P DGFRA H
e t
2345 R25L*, A167T*, R681H *, C708R, R1504Q* S361R, T474M,S
478P
MP D
GIST, idiopathic hyperosinophilic
syndrome
t
SFRP 4 H
e t
STEAP 2 H o m
TCF3 H
345 P 320T, R340K
489 F17C*, R456Q *, M475I
653 P 479L pre B-ALL
P LAG1 H
e t
P ML H
e t
P MS2 H o m
P OU6F2 H o m
PP ARGC1A H
o
500 S443R salivary aden o- ma , pleomorphic adenoma
828 S722G AP L
861 P 470S*, T485K *, K541E
691 P 191L
797 G482S
colorectal, endometri- al, ovarian, medullo b- lastoma, glioma
e t
TEK H o m
TFEB H
e t
TFRC H
e t
THBS4 H o m
TMP RSS2 H
e t
1123 I148T*, Q346P
475 V130M renal (chil d- hood) epitheli o- id
760 G142S NHL
1538 I192T, I598T, S1055G
491 V160M prostate
m
P TP N11 H
e t
P TP N21 H
e
592 S189A, Y197st (TAT-
>TAG)
1173 L385F, V936A
JMML, AML, MDS
TNC H o m
TNFRSF10A H
o m
2200 V295M*, Q539R, V605I, E2008Q
467 H141R, R209T,
R441K
glioma, lung/colon/breas t cancer![]()
t
P VRL4 H
e
509 F53L
TNFRSF17 H o m
183 N81S intestinal T-cell lymphoma
IJSER © 2012
Inte rnatio nal Jo urnal o f Sc ie ntific & Eng inee ring Re se arc h Vo lume 3, Issue 3 , Marc h-2012 8
![]()
ISSN 2229-5518

TRAF3 H o m
TSC1 H
e t
USP 6 H
e t
WISP 3 H
e t
568 M129T
365 M322T
234 Y162H, W475R, Y484H
331 Q34H,
E100K, E141K
aneurysmal bone cysts
colon cancer hamarto ma, renal cancer
cancer-susceptibility patients or direct sequencing the tumor samples and paired germline genomes.
In summary, we developed targeted exome capture sequen c- ing technology to characterize the whole-exome of human genome and applied to a high-cancer-susceptible patient. We showed that the truncations of CARD8, MAGEE2, ANAPC1, GPR1, ASCC3, MAP2K3 and PTPN11 be an important reasons for high-cancer-susceptiblity. The non-synonymous mutations in 132 cancer-associated genes, in which most of them have not been reported as germline variations in tumors, may pos i- tively or negatively contribute to cancer development. This exome sequencing technology makes it possible for routine dissection of important genes for carcinogenesis and indi vidu-
ALCL, anaplastic large-cell lymphoma; ALL, acute lymphocytic leukemia; AML, acute myelogenous leukemia; APL, acute promyelocytic leukemia; B -ALL, B-cell acute lymphocytic leukaemia; CMML, chronic myelomonocytic leukemia; CNS, central nervous system; DLBL, diffuse large B-cell lymphoma; DLCL, diffuse large- cell lymphoma; GIST, gastrointestinal stromal tumour; JMML, juvenile myelomo- nocytic leukemia; MDS, myelodysplastic syndrome; MLCLS, mediastinal large cell lymphoma with sclerosis; MM, multiple myeloma; MPD, Myeloproliferative disord- er; NHL, non-Hodgkin lymphoma; NSCLC, non small cell lung cancer; pre-B All, pre-B-cell acute lymphoblastic leukaemia; SKC, skin squamous cell; T-PLL, T cell prolymphocytic leukaemia. *listed as heterozyous mutation.
The Cancer Genome Atlas project is currently the central task of genome-related research. It remains largely unknown how germline mutations in global contribute to cancer- susceptibility, although it is well known some germline muta- tions in a special gene would cause human cancers (e.g., mu- taions in pRB gene leads to retinoblastoma in children). The major challenge is to develop a high throughput and cost- effective techniques for genome sequencing. Supported with extensive bioinformatic assays, a US group and us have ind e- pendently developed cost-effective targeted capture exome sequencing technology to routinely reveal the genetic varia- tions of individuals. However, to our knowledge, the whole exome sequencing on high-cancer-susceptible patient has not yet been studied. In this study, we independently developed a similar technology for the whole exome sequencing. As a pilot study, we showed that homozygous mutations of CARD8 may contribute to the high-cancer-susceptibility in a patient, who underwent three high mortality cancers (breast cancer, gallbladder cancer and lung cancer) in the last three decades. CARD8 was reported to inhibit apoptosis and caspase activa- tion induced by Apaf-1/caspase-9-dependent stimili; howev- er, it was also showed to induce apoptosis in certain cells. It is unclear how the loss of CARD8 contributes the high -cancer- susceptibility in this patient. The mutations in other genes, such as genes on RAS-MARK signaling pathway, may also play important roles in high -cancer-susceptibility. However, as some mutations may neutralize or antagonize the other mu- tations, the exact roles of these mutations are very complicated in the patient. For example, the truncation of MAGEE2 and PTPN11 may neutralize the mutations of tyrosine kinases and GPRs. The roles of these mutations in cancer-susceptibility would be further investigated by identification of more high -
alized medicine, as the total cost is just less than US$10,000 per
sample. The targeted exome capture sequencing would be a new era of individualized cancer therapy.
This study was supported in partial by Shenzhen -Hong Kong Collaborative Research Grant of Shenzhen Science and Tech- nology Bureau (08DF-23, to ML He and Y He) and Research Grant Council, The Government of Hong Kong Special Ad- ministration Region (CUHK4428/ 06M, to MLHe).
No conflicts of interest.
[1] Morrow PK, Hortobagyi GN. Management of breast cancer in the genome era. Annu Rev Med. 2009;60:153–165.
[2] Kawashima M, Fuwa N, Myojin M, Nakamura K, Toita T, Saijo S, Hayashi N, Ohnishi H, Shikama N, Kano M, Yamamoto M. A multi-institutional survey of the effectiveness of chemotherapy combined with radiotherapy for patients with naso- pharyngeal carcinoma. JpnJ ClinOncol. 2004;34:569–583.
[3] Gutierrez ME, Kummar S, Giaccone G. Next generation oncology drug devel- opment: opportunities and challenges. Nat Rev ClinOncol. 2009;6:259–265.
[4] Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378.
[5] Taylor BC, Yuan JM, Shamliyan TA, Shaukat A, Kane RL, Wilt TJ. Clinical out- comes in adults withchronic hepatitis B inassociation withpatient and viral charac- teristics: A systematic review of evidence. Hepatology. 2009;49:S85–95.
[6] Diller L, Chow EJ, Gurney JG, Hudson MM, Kadin-Lottick NS, Kawashima TI, Leisenring WM, Meacham LR, Mertens AC, Mulrooney DA, Oeffinger KC, Packer RJ, RobisonLL, Sklar CA. Chronic disease inthe Childhood CancerSurvivor Study cohort: a review of published findings. J ClinOncol. 2009;27:2339–2355.
[7] Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature.
2009;461:272–276.
[8] Wang J, Wang W, Li R, Li Y, Tian G, GoodmanL, FanW, Zhang J, Li J, Zhang J, Guo Y, Feng B, Li H, Lu Y, Fang X, Liang H, Du Z, Li D, Zhao Y, Hu Y, Yang Z,
IJSER © 2012
Inte rnatio nal Jo urnal o f Sc ie ntific & Eng inee ring Re se arc h Vo lume 3, Issue 3 , Marc h-2012 9
ISSN 2229-5518
Zheng H, HellmannI, Inouye M, Pool J, Yi X, Zhao J, Duan J, Zhou Y, QinJ, Ma L, Li G, Yang Z, Zhang G, Yang B, Yu C, Liang F, Li W, Li S, Li D, Ni P, RuanJ, Li Q, Zhu H, Liu D, Lu Z, Li N, Guo G, Zhang J, Ye J, Fang L, Hao Q, Chen Q, Liang Y, Su Y, SanA, Ping C, Yang S, ChenF, Li L, Zhou K, Zheng H, Ren Y, Yang L, Gao Y, Yang G, Li Z, Feng X, Kristiansen K, Wong GK, Nielsen R, Durbin R, Bolund L, Zhang X, Li S, Yang H, Wang J. The diploid genome sequence of an Asian individ- ual. Nature. 2008;456:60–65.
[9] Schaefer A, Jung M, Kristiansen G, Lein M, Schrader M, Miller K, Erbersdobler A, Stephan C, Jung K. [MicroRNA in uro-oncology : New hope for the diagnosis and treatment of tumors?] Urologe A. 2009
[10] Mutesa L, Pierquin G, Janin N, Segers K, Thomee C, Provenzi M, Bours V. Germline PTPN11 missense mutation in a case of Noonan syndrome associated withmediastinal and retroperitoneal neuroblastic tumors. Cancer Genet Cytogenet.
2008;182:40–42.
[11] Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An over- view of the MAGE gene family with the identificationof all humanmembers of the family. CancerRes. 2001;61:5544–5551.
[12] Razmara M, Srinivasula SM, Wang L, Poyet JL, Geddes BJ, DiStefano PS, Bertin J, Alnemri ES. CARD-8 protein, a new CARD family member that regulates cas- pase-1 activationand apoptosis. J Biol Chem. 2002;277:13952–13958.
[13] Heichman KA, Roberts JM. The yeast CDC16 and CDC27 genes restrict DNA
replicationto once per cell cycle. Cell. 1996;85:39–48.
[14] Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27HS colocalizes with CDC16HS to the centrosome and mitotic spindle and is essential forthe metaphase to anaphase transition. Cell. 1995;81:261–268.
[15] Ahuja A, Ying M, Evans R, King W, Metreweli C. The application of ultra- sound criteria for malignancy in differentiating tuberculous cervical adenitis from metastatic nasopharyn-geal carcinoma. Clin Radiol. 1995;50:391–395.
[16] Jorgensen PM, Brundell E, Starborg M, Hoog C. A subunit of the anaphase- promoting complex is a centromere-associated protein in mammalian cells. Mol Cell Biol. 1998;18:468–476.
[17] Rosenbaum DM, RasmussenSG, Kobilka BK. The structure and function of G- protein-coupled receptors. Nature. 2009;459:356–363.
[18] De Meyts P, Gauguin L, Svendsen AM, Sarhan M, Knudsen L, Nohr J, Kise- lyov VV. Structural basis of allosteric ligand-receptor interactions in the insu- lin/relaxin peptide family: implications for other receptor tyrosine kinases and G- protein-coupled receptors. Ann N YAcad Sci. 2009;1160:45–53.
[19] PathanN, Marusawa H, Krajewska M, Matsuzawa S, Kim H, Okada K, Torii S, Kitada S, Krajewski S, Welsh K, Pio F, Godzik A, Reed JC. TUCAN, anantiapoptot- ic caspase-associated recruitment domain family proteinoverex-pressed in cancer. J Biol Chem. 2001;276:32220–32229.
———— ——— ——— ——— ———
Soniyapriyadharishni.A.K. is currently pursuing Ph.D program in Bioinfor- maticts in Bharath University, India,
Dr.M.Sridhar,D.Sc.,A.Sc., is currently the Director A&P inBharath Universi- ty, India
Dr.M.Rajani,D.Sc.,is the Director R&D in Bharath University, India.
IJSER © 2012