International Journal of Scientific & Engineering Research, Volume 6, Issue 3, March-2015 87
ISSN 2229-5518
Molecular detection of ciprofloxacine resistance of Salmonella typhi
Hazim A. N. Alhadrawi*, Mahdi Hussain Al-Ammar , Haider Laitef
Abstract—The study was conducted to susceptibility to antibiotics and molecular level analysis of the cause of reduced sensitivity of Salmonella typhi isolates from patients in Babylon . Out of 50 blood cultures obtained during the study, 12 (14.0%) showed positive blood cultures were due to S. typhi and rests were mostly of S. paratyphi A. The prevalence was highest between the age group 3 - 15 year.. Among all S. typhi isolates, 41.4% were sensitive to ampicillin, cotrimoxazol and chloramphenicol, respectively. All isolates were sensitive to ceftriaxon and ceftazidim; 5 isolates were ciprofloxacin resistant, others were moderate to highly sensitive; whereas, only 2.2% isolates were sensitive and almost all (97.8%) were found resistant to nalidixic acid. The E-strip test among isolates showed the MIC value nearer to the sensitive between 0.125-0.5 and rest other isolates showed from > 2.0 μg/ml to very highly resistant. VNTR pattern of all ciprofloxacine resistant S. typhi was also same. Restriction fragment analysis of gyrase‐A gene indicated point mutations in different loci that bear the cause of being resistant to ciprofloxacin.
Index Terms— Salmonella entericaserovarParatyphi A, DNA gyrase, Typhoid fever, Salmonella typhi, Fluoroquinolone resistance.
1 INTRODUCTION
—————————— ——————————
Typhoid fever is a major cause of morbidity and mortality with anestimated global incidence in world.1 Salmonella enterica- serovar Typhi (S. Typhi) is responsible for the majority of cas- esfollowed by S. Paratyphi A(1) . In the lasttime, the world- wideemergence of multidrug-resistant strains of Salmonella has led tovirtual withdrawal of chloramphenicol and its replacement withfluoroquinolones and third-generation cephalosporins(.2,3) Nalidixic-acid-resistant strains exhibiting reduced susceptibilityto ciprofloxacin (MICs 0.125–1 mg/L) (4).Clinical treatment fail- ures after the administration of ciprofloxacinand other fluoro- quinolones to patients with typhoid feverattributable to these strains have been reported.(4,5).
Recent reports ofinfections because of strains of S. Paratyphi A with high-levelresistance to fluoroquinolones are therefore par- ticularlyworrying.(5,6) The targets of fluoroquinolones are the two enzymes.
DNA gyrase and topoisomerase IV, whose subunits are encod- edrespectively by gyrA and gyrB and the parC and parE genes. Thealteration caused by single point mutations within the quin- oloneresistance-determining region (QRDR) of the DNA gyrase subunitgyrA gene leads to quinolone resistance.(7) In Salmonella, the most commonresidues associated with mutation leading to quinolone resistancehave been Ser-83 and Asp-87 in the gyrA gene, either alone ortogether.(4,7)Additional mutations may be required to attainhigh-level fluoroquinolone resistance.(10,8,9).
————————————————
• Author name is Hazim A. N. Alhadrawi biologist. phd dgree in Microbi- ology. University of Kufa, Iraq. E-mail: hazim.alhadrawi@uokufa.edu.iq
• Co-Author name is Mahdi Hussain Al-Ammar . phd dgree. University
of Kufa, Iraq. E-mail: Dr.mahdi65@yahoo.com
Haider Laitef master degree . University of Kufa, Iraq. E-mail:
haider.laitef@uokufa.edu.iq
Complete fluoroquinoloneresistance in the Enterobacteriaceae usually results from twoor more point mutations within the QRDRs of the DNA gyraseand topoisomerase IV genes(9,10)
To our knowledge this is the first report in Iraq of molecular- characterization of S. Typhi showing a full fluoroquinolonere- sistance phenotype causing enteric fever. The molecularcharacter- istics of ciprofloxacin-resistant isolates of S. Typhi were com- pared with those of strains fully susceptibleto ciprofloxacin and with reduced susceptibility to ciprofloxacin.(11,12).
2.2 Materials and Methods
2.1. Bacterial Strains
A total of 12 isolates, which included S. Typhi strains isolated from blood cultures of patients suffering from enteric fever, were studied .The blood samples were collected from unvaccinated patients who were clinically diagnosed and admitted to Babylon hospital. Blood cultures were made using standard methodology on Blood-Agar, Chocolate-Agar and MacConkey-Agar medi- um,these included five ciprofloxacin-resistant strains, were iden- tified by standard biochemical tests and agglutination using spe- cific antisera (Murex com.).stored at -20 °C.. The isolates of S. typhi were confirmed by standard biochemical (API-20E system, BioMeriux,) (13)c compared withstandard results ofS. Typhi was used as control.
2.2. Antimicrobial Susceptibility Testing
Antibiotic sensitivity and minimum inhibitory concentration for fluoroquinolone (ciprofloxacin) were performed on Mueller Hinton agar using disc diffusionmethod in accordance with Na- tional Committee forClinical Laboratory standards (NCCLS) (14).The antibiotics tested were gentamicin, amika- cin,piperacillin, ciprofloxacin, ceftazidime, piperacil- lin,ceftriaxone and ceftizoxime (Hi-media Laboratories). MICs of ciprofloxacin were determined by agar dilution and final analysis was done using an E-test kit.
IJSER © 2015 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 6, Issue 3, March-2015 88
ISSN 2229-5518
2.3. S Typhi DNA Extraction
DNA was extracted from pure culture of Salmonella typhi on MacConkey-agar plate by heat-lysis method. Molecular typing was done by PCR using primers as described by Liu et al (1995). The PCR amplified product of gyraseA gene was confirmed by running it through gelelectrophoresis. (15,16).
3. RESULTS AND DISCUSSION
Out of the 50 blood cultures obtained during the study period,
12 (14.0%) yielded significant growth were S. typhi. The isola- tion rate from boys and girls was apparently similar .The present
laboratory‐based study, showed the prevalence of S. typhi
among the age group between 2 and 10 years. . In addition, the isolation rate was highest in the summer and monsoon season, with peaks in September and was relatively low from November to March. Among all Salmonella typhi isolates studied, 42.6% of the total isolates were sensitive to ampicillin, whereas 57.4% resistant; 42% of the total isolates were sensitive to cotrimoxazol, whereas 57.4% are resistant. On the other hand, all isolates were sensitive to ceftriaxon and ceftazidim; 41.4% of the total isolates were sensitive to chloramphenicol and 58.6% are resistant; 5 iso- lates collected were ciprofloxacin resistant, others were moderate to highly sensitive; however, only 2.2% isolates were nalidixic acid sensitive and almost all (97.8%) were found resistant (Fig.
1).
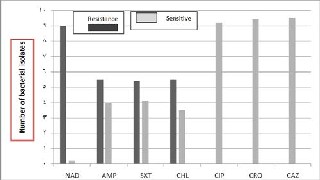
Antibiotics
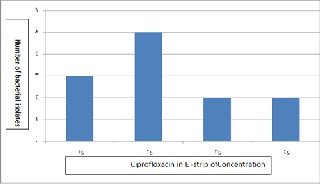
Fig. 1. Antibiogram profile of Salmonella typhi to different antibiotics
4
2.0 μg/ml, respectively.).
Present study shows an increase in the reduced fluoroquino- lone susceptibility from 3.9 to 23.5% among all the Salmonella serovar typhi . Among all 12 S. typhi isolates studied, E-strip test was performed for isolates of which 3 showed the MIC value nearer to the very sensitive (< 0.125 μg/ml), 5 showed between
0.125 to 0.5 μg/ml, 2 showed between 0.5 to 2.0 μg/ml and rest other 2 isolates showed from > 2.0 μg/ml to vary highly resistant e.g. 512 μg/mlfigure .2.
All the ciprofloxacine resistant isolates were also highly re- sistant to ampicillin (> 256 μg/ml), cotrimoxazole (> 32 μg/ml), chloramphenicol (> 256 μg/ml), ciprofloxacin (> 32 μg/ml) and nalidixic acid (> 256 μg/ml), and were susceptible to ceftriaxone (0.094 μg/ml) according to their MIC of respective antibiotics. All isolates were found to be identical by Api 20E) (Fig. 3).
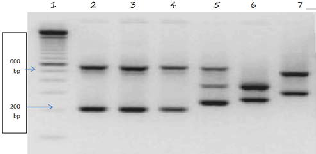 6
6
Fig. 3. VNTR pattern of ciprofloxacin resistant S. typhi isolates with that of com- parative sensitive isolates (Lane 1: 100 bp marker; Lane 2,3,4: Ciprofloxacin resistant isolates, Lane 5,7: Ciprofloxacin sensitive isolates.
Transferable, mutational resistance, and clonal spread are reasons for the rapidly increased quinolone resistance in Sal- monella typhi isolates,(5) As far as we know, however, transfer- able fluoroquinolone resistance appears to be rare in bacteria in vivo. Thus, either clonal spread or resistance due to muta- tions in chromosomal genes remains the potential mechanism accounting for the high level of reduced fluoroquinolone sus- ceptibility. In Clonal spread as a major contributing factor was excluded by identification of 5 serotypes among the quino- lone-resistant isolates. In addition, some of these serotypes contained different antimicrobial resistance patterns along with their different VNTR patterns. Based on these data, it could bewe can conclude that the reduced fluoroquinolone susceptibility of Salmonella typhi in Iraq primarily involves mutations in the chromosomal genes. This concept is identify with our experiment and finally proved by sequencing data where all Salmonella typhiisolates with reduced fluoroquino- lone susceptibility were analyzed in gel electrophoresis have shown point mutation leading to nucleotides change in their QRDR of the gyraseA gene.
Taq-DNA polymerase successfully amplified the 195 bps gy-
raseA gene from the genomic DNA of ciprofloxacin sensitive and
ciprofloxacin resistant Salmonella serover typhi using primer of
gyraseA (Fig. 4).
Fig. 2. Status of susceptibility of S. typhi isolates to Fluoroquinolone. (S1 is <
0.125, S2 is in between 0.125 and 0.5, S3 is in between 0.5 and 2.0 and S4 is >
IJSER © 2015 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 6, Issue 3, March-2015 89
ISSN 2229-5518
Fig. 4. Amplified gyraseA gene using PCR.
The increase in the incidence of fluoroquinolone resistance in Salmonella and other enteric bacteria, especially Campylobacter sp. and E. coli(6-16,), drives a situation that impede the effec- tiveness of this antimicrobial group. The significantly common multi drug resistance observed here among the fluoroquino- lone-resistant Salmonella compared with the susceptible popula- tion (47.4% vs. 11.5%) is also a matter of concern. This finding suggests that the abrupt use of fluoroquinolone for multidrug resistant Salmonella give proven of whether the same could hap- pen to other bacterial species. Collectively, these data indicate that safe use of the fluoroquinolone antimicrobial group is war- ranted to prevent further development of resistance and to pre- serve the usefulness of this valuable drug.(17,18,19).
In conclusion, it has been shown that reduced susceptibility of Salmonella to the fluoroquinolone group was significantly asso- ciated with multidrug resistance. Moreover, all quino- lone-resistant Salmonella isolates had undergone a point mutation in the QRDR of the gyrA gene. In contrast to previous reports on quinolone resistance in a specific clone or in a few Salmonella serotypes, the reduced fluoroquinolone susceptibility of our iso- lates was nonclonal(20).These data give more evidence of the rapid spread of multidrug-resistant pathogens from one country to another. The result of antimicrobial resistant pathogen in any city of the world may have universal finding and is therefore a general concern.
In Gram-negative bacteriathe primary target of fluoroquin- olones is gyrase rather thantopoisomerase IV, hence gyrAmuta- tions precede those of Styphi. Since eachmutation in gyrAwas associated with differentciprofloxacin MICs, further studies on other resistance mechanisms,such as alterations in membrane permeability and changesin efflux and influx, are required to evaluate the contribution ofparCmutations to fluoroquinolone resistance in S. typhi and are presently under investigation(1).
This study suggests that isolates with reduced susceptibility to- fluoroquinolones might be important in clinical development ofresistance as they could become highly resistant upon sequen- tialacquisition of resistance. Double mutations in gyrA, along with asingle mutation in parC, have also been reported in in vitroselected ciprofloxacin-resistant mutants of S. Paratyphi A,strongly suggesting that such triple mutation is important for thedevelopment of high-level fluoroquinolone resistance(3).
All ciprofloxacin-resistant S. typhi isolates demonstratedan identical PFGE pattern and mutations in DNA gyraseand topoi- somerase IV as did the S. Paratyphi A isolates. Thepatients in- fected with these resistant isolates did not give a historyof prior treatment with fluoroquinolones. This is the first report in Iraq
country suggesting the spread and the infection by a circulating resistantstrain rather than the emergence of resistance during treatment. There is widespread availability and uncontrolled use of antibioticsincluding quinolones. Therefore, there is selective pressure on alarge bacterial population of endemic Salmonella spp. Reducingthe exposure to fluoroquinolones would definitely lessen thelikelihood of selecting mutants. As isolates with re- ducedsusceptibility to fluoroquinolones could become highly resistantupon sequential accumulation of mutations in topoiso- merasegenes, the use of fluoroquinolones as first-line drugs for- management of enteric fever in areas where these strains areendemic, therefore, requires urgent review.Continuous surveil- lance of the plasmid and chromosome ofS. typhi is essential to alter treatmentstrategies aimed at maintaining the useful life of the fewremaining antimicrobials available to treat enteric fe- ver(4,5).
ACKNOWLEDGMENT
Praise to the mighty "Allah" (SWT) who gave me health, strength, and facilitated the ways for me to accomplish this work. It is a pleasure to express my deep appreciation to my Friend for the scientific guidance and support. I would like to thank all members of Biology Department and The Deanery of College of Sciences-Kufa University for their kind cooperation. My thanks to the staff members of Bacteriology laboratories in Al-Sadder Medical city in Najaf for facilitating the collection of specimens.
REFERENCES
[1] Crump JA, Lubsy SP, Mintz ED. The global burden of enteric fever.
Bull World Health Organ 2004; 82: 346–53.
[2] Bhan MK, Bhal R, Bhatnagar S. Typhoid and paratyphoid fever.
Lancet 2005; 366: 749–62.
[3] Kariuki S, Gilks C, Revathi G et al. Genotypic analysis of multi- drug-resistant Salmonella enterica serovar Typhi, Kenya. Emerg In- fect Dis 2000; 6: 649–51.
[4] Smith KE, JM Besser, CW Hedberg, FT Leano, JB Bender, JH Wicklund. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N. Engl. J. Med. 1999, 340: 1525-1532.
[5] Sáenz Y, M Zarazaga, M Lantero, MJ Gastañares, F Baquero, C Torres. Antibiotic resistance in Campylobacter strains isolated from animals, foods, and humans in Spain in 1997-1998. Antimicrob Agents Chemother 2000,44: 267-271.
[6] Reyna F, M Huesca, V Gonzalez and L Y Fuchs. Salmonella typhi- murium gyrA mutations associated with fluoroquinolone resistance. Antimicrob Agents Chemother 1995,39: 1621-1623.
[7] Prats G, B Mirelis, T Llovet, C Muñoz, E Miró, F Navarro. Antibi- otic resistance trends in enteropathogenic bacteria isolated in
1985-1987 and 1995-1998 in Barcelona. Antimicrob Agents
Chemother2000,44: 1140-1145.
[8] Adachi T, Sagara H, Hirose K et al. Fluoroquinolone-resistant Sal- monella Paratyphi A. Emerg Infect Dis 2005; 11: 172–4.
[9] Piddock LJ, Ricci V, McLaren I et al. Role of mutation in gyrA and parC genes in nalidixic-acid-resistant salmonella serotypes isolated from animals in the United Kingdom. J Antimicrob Chemother
1998; 41: 635–41.
IJSER © 2015 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 6, Issue 3, March-2015 90
ISSN 2229-5518
[10] Cloeckaert A, Chaslus-Dancla E. Mechanism of quinolone re-
sistance in Salmonella. Vet Res 2001; 32: 291–300.
[11] Cebrian L, Sirvent E, Rodriguez Diaz JC. Characterization of Sal- monella spp. mutants produced by exposure to various fluoroquin- olones. Int J Antimicrob Agents 2003; 22: 134–9.
[12] Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents 2005; 25: 358–73.
[13] Ackers ML, Puhr ND, Tauxe RV et al.Laboratory based surveil- lance of Salmonella serotype Typhi infections in U| States- antimicrobial resistance on the rise. JAMA 2000; 283: 2668-73.
[14] National Committee for Clinical Laboratory Standards.Seventh
Edition: Approved Standard M2–A7. NCCLS, Wayne, USA, 2002.
[15] Ausubel FM, Brent R, Kingston RE (eds). Current Protocols in
Molecular Biology. New York: John Wiley & Sons, 1996.
[16] Nair S, Unnikrishan M, Turner K et al. Molecular analysis of fluo- roquinolone-resistant Salmonella Paratyphi A isolate, India. Emerg Infect Dis 2006; 12: 489–91.
[17] Martínez-Martínez L, A Pascual, GA Jacoby 1998. Quinolone re- sistance from a transferable plasmid. Lancet 351: 797-799.
[18] Kariuki S, Revathi G, Muyodi J et al.Characterization of multire- sistant typhoid outbreaks in Kenya. J Clin Microbiol 2004; 42:
1477–82.
[19] Renuka K, Seema S, Das B et al. High-level ciprofloxacin re- sistance in Salmonella enterica serotype Typhi in India. Letter. J Med Microbiol 2005; 54: 999–1000.
[20] Fluit AC. Towards more virulent and antibiotic-resistant Salmonel- la. FEMS Imm Med Microbiol 2005; 43: 1–11.
IJSER © 2015 http://www.ijser.org



