
M.P.E. Shaekh, A. Mondol, M.M. Islam, A.S. Kabir, M.A. Saleh, M. Salah Uddin, K.M.F. Hoque, A.E. Ekram*.
Abstract— Five single bacterial colonies were isolated aseptically, from which one colony showed antagonistic effect against Agrobacterium tumefaciens, E.coli BL21, Enterobacter sp. and Pseudomonas aeruginosa with 14mm, 14mm, 10mm and 13mm zones of inhibition respectively and was characterized of the antagonistic bacterium. The optimum culture condition of the isolate was at pH 8.5. The bacteria showed the ability of utilizing arabinose, sucrose, maltose and fructose as carbon sources and viable cell count was 459×107 CFU/ml. The bacterium was resistant to amoxicillin, penicillin and vancomycin and the Minimum Inhibitory
Concentration (MIC) value against vancomycin was 50µg/ml. The molecular weight of isolated bacterial genomic DNA was above 10 kb with not much smearing. Current research was mainly focused on searching for antagonistic bacteria by morphological, physiological, biochemical tests, which may be used to control emerging pathogens.
Index Terms— Antagonistic effect, Penaeus monodon, Minimum Inhibitory Concentration (MIC), genomic DNA.
—————————— ——————————

M
icroorganisms display interesting competitive mechanisms, of which antagonism has been commonly referenced [1,2]. Microbial antagonism is biological phenomenon in which certain microorganisms of the normal microbiota that suppresses the growth of other microorganisms through competition for nutrients and the secretion of inhibitory substances. Some microorganisms can inhibit other microorganisms or reduce their growth in medium thanks to their metabolites, indirect (by changing pH, osmotic pressure and surface tension) or direct (by producing toxic component, antimicrobial component, bacteriocin, antibiotic etc.), this situation is called as antagonistic relation. Microorganisms are not only the cause of infections; they can also produce organic substances that
M.P.E. Shaekh, M.M. Islam, A.S. Kabir are MS students in Department of Genetic Engineering and Biotechnology and A. Mondol is MS student in Department of Zoology, University of Rajshahi, Rajshahi-6205, Bangladesh.
M.A. Saleh, M. Salah Uddin, A.E. Ekram are Assistant Professor and
K.M.F. Hoque is associate Professor in Department of Genetic Engineering
and Biotechnology, University of Rajshahi, Rajshahi-6205, Bangladesh.
can cure infections [3]. The first marine bacterium based antibiotic was characterized in 1966 [4]. As the primary role of antimicrobial activity can be to antagonize competitors, bacteria may also produce antimicrobial compounds when they sense the presence of competing organisms [5]. Many microorganisms contain substances that have antimicrobial, antiviral, anticoagulant and cardio active properties. A few of these substances have unique chemical structures that are unlike any other compounds, may serve as leads to the discovery of new drugs. The Streptomyces sp. isolated from the surface of a jellyfish produced two bicyclic peptides, salinamides A and B [6]. Likewise, Bacillus isolated from a marine worm produced a novel cyclic decapeptide antibiotic lolatin B [7].
The bacterium Pelagiobacter variabilis isolated from the seaweed Pocockiella variegata produced phenazine antibiotics, pelagiomicins [8]. Microbial competition for limiting natural resources within a community is thought to be an important selective force that promotes biosynthesis of antimicrobial compounds [9]. Biological control using antagonistic bacteria has been reported as an attractive alternative due to their ability to antagonize the pathogen by different modes of action.
Bacteria in aquatic ecosystems might produce antimicrobial substances inhibiting the growth of other microorganisms [10]. Fish are currently protected from infectious diseases by vaccination or chemotherapeutic treatment. However, due to extensive use of the chemotherapeutic agents, the occurrence of antimicrobial- resistance among pathogens and the associated environmental problems have been well documented [11, 12, 13]. Fish in production facilities are exposed to stressful conditions, diseases and deteriorating environmental conditions, all of which can result in serious economic losses [14]. Therefore, several alternative strategies including the use of antagonistic bacteria have been proposed. More than half of 86 isolates from the Southern Californian Bight showed antagonistic activity and especially bacteria from marine snow displayed strong antagonistic activities towards other bacteria [15].
In Scottish coastal waters 35% of surface-associated bacteria of various seaweed and invertebrate species were shown to produce antimicrobial compounds [16]. The marine environment harbours a wide range of microbes capable of exhibiting bacteriolytic and antibiotic activity and the primary role of the antibiotic substances could be attributed to ecological competition, the beneficial associations between associated bacteria [17]. Enterobacter sakazakii has been associated with powdered infant formula outbreaks which caused high mortality rate illnesses in infants, Enterobacter infections can include bacteremia, lower respiratory tract infections, skin and soft-tissue infections, urinary tract infections (UTIs), endocarditis, intra-abdominal infections, septic arthritis, osteomyelitis, CNS infections and ophthalmic infections.
Agrobacterium tumefaciens, the cause of the economically important disease, crown gall, is cosmopolitan in distribution, affecting dicotyledonous plants in more than
60 different plant families. Pseudomonas aeruginosa is a
gram-negative bacterium that continues to be a major
cause of opportunistic nosoco-mial infections, causing around 9–10% of hospital infections. It is also the dominant cause of chronic lung infections contributing to the death of patients with cystic fibrosis. A major reason for its prominence as a pathogen is its high intrinsic resistance to antibiotics, such that even for the most recent antibiotics, a modest change in susceptibility can thwart their effectiveness. The increasing incidence of antibiotic resistance among bacterial pathogens and emerging new diseases are posing great challenges to humans. Given the widespread misuse and over prescription of antibiotics by the medical community, the antibiotics available today are rapidly becoming less and less effective in the face of
emerging multi drug resistant pathogens of clinical concern [17]. That is why, antagonistic bacteria from different sources including fish may be used as biocontrol agent or probiotics or alternative to antiobiotics.
For the experimental purpose, cuticle from black tiger shrimps was used as source of inocula. Juvenile black tiger shrimps (P. monodon) were collected from commercial farms in Sriula, Satkhira, Bangladesh after ensuring its standard and healthy condition. Aseptic environment was maintained during collecting, transporting and storing the sample shrimps.
Isolation and identification of microorganisms from natural resources provide a unique source for acquiring pure cultures with potential commercial utility [18]. In the present study, cuticle was separated from Penaeus monodon and crushed in saline solution aseptically. Then sample of the crushed material was filtered and finally inoculated into LB medium. The inoculated conical flask was incubated in a rotary shaker at 160 rpm, 370C for 24h. That bacterial culture was used as source material to collect bacterial colony. A single bacterial colony was isolated aseptically by plating the bacterial culture of the crushed cuticle of Penaeus monodon onto an agar solidified LB medium. The plates were incubated at 37°C for 24 hours and bacterial colonies were found to grow on the medium. The Bacterial isolates were screened on nutrient agar supplemented media with 10gm/l peptone, 5gm/l Yeast Extract, and 20gm/l Agar powder. Screening was done for a period of 18hours. Species identification was performed using standard microbiological and biochemical methods.
Pure cultures of Agrobacterium sp., E.coli BL21, Enterobacter sp. and Pseudomonas aeruginosa were obtained from the Microbiology Laboratory, Department of Genetic Engineering and Biotechnology, University of Rajshahi, for the selection of antagonistic bacteria.
The stock of Agrobacterium sp., Bacillus subtilis, E.coli BL21, Enterobacter sp. and Pseudomonas aeruginosa were taken out of -200C and revived in LB medium. Culture was incubated at 370C for overnight. These suspensions were used as inoculums. On the other hand, sample bacterium was also sub-cultured.
Before disc preparation, subculture was centrifuged to get culture medium without bacteria. So we had following three sample:
Subculture of bacteria
LB medium
Cell free supernatant
Disk diffusion method was used in the current experiment to evaluate the antagonistic activity of selected bacterium. The discs (6 diameters) were made by punching the Whatman No.1 filter paper with the help of punch machine. These discs were taken into the screw capped tube and sterilized in an autoclave machine at 1210C for 20 minutes to ensure sterilization. The paper discs were soaked with 20µl concentration of bacteria with the help of micropipette and keep them at laminar air flow hood for dryness (5-10 minutes). The disks contain sample, normal medium as well as the centrifuged culture medium disks were introduced on the upper layer of the seeded agar plate by sterile forceps. Then the plates were incubated overnight at 370C.
Morphological observation of the bacteria was performed through microscopic observation. For motility test medium 10.0g/l, NaCl 5.0g/l, agar 4.0g/l was used and a sterile needle was used to pick a well isolated colony and stabled the medium to within 1 cm of the bottom of the test tubes. A positive motility test was indicated by a growth area extending away from the line of incubation. A semisolid agar called Sulfide-Indole-Motility medium (or SIM medium) is inoculated with bacteria to test for hydrogen sulfide, indole and motility of the organism. The indole test screens for the ability of an organism to degrade the amino acid tryptophan and produce indole. If hydrogen sulfide is present, it will react with the sodium thiosulfate in the medium and the indicator, ferric ammonium citrate, to produce ferrous sulfide which falls out of solution as a blackish precipitate. The presence of hydrogen sulfide typically means that the bacteria produce the enzyme cysteine desulfanase which breaks up the cysteine in the medium into, among other components, hydrogen sulfide.
Basal salt media collected with methyl red was used for
decolonization studies. The citrate test was done to screen out a bacterial isolate for the ability to utilize citrate as its carbon and energy source. Catalase production and activity was detected by adding the substrate H2 O2 to an appropriately incubated (18 to 24 hour) tryptic soy agar
slant culture. MacConkey agar was used for the isolation of gram-positive bacteria and the differentiation of lactose fermenting from lactose non-fermenting Gram-positive bacteria. The isolated bacterial strains were grown overnight in nutrient broths that were placed in the shaker at 370C temperature and 120rpm for the antibiotic sensitivity test. A serial dilution technique was made for the test respective. The number of living cells per ml of solution was observed by viable cell count through serial dilutions techniques. Determination of Minimum Inhibitory Concentration (MIC) of vancomycin was done by broth tube dilution method. Isolation of bacterial genomic DNA was performed with 1kb DNA ladder. In order to find out the ability of the isolates to utilize different carbohydrates, the cultures were inoculated in MS medium containing different carbohydrates viz. arabinose, sucrose, lactose, maltose, galactose, xylose and fructose.
Cuticle was dissected from Penaeus monodon and crushed in saline solution aseptically. Then sample of the crushed material was filtered and finally inoculated into LB medium. The inoculated conical flask was incubated in a rotary shaker at 160 rpm, 370C for 24h. That LB culture was used as source material to isolate bacterial colony. The antagonistic bacterial colony was isolated through disc diffusion assay method [19], against four bacterial strains such as Agrobacterium tumefaciens, E.coli BL21, Enterobacter sp. and Pseudomonas aeruginosa. Only the isolated bacterium showed zone of inhibition but the disk containing only LB medium and cell free supernatant showed no zone of inhibition. This result indicated that the zone of inhibition was resulted due to antagonistic activity of the bacterium and there was no effect in case of only LB medium and cell free supernatant. The bacterium showed 14mm, 14mm, 10mm and 13mm zone of inhibition against Agrobacterium tumefaciens, E.coli BL21, Enterobacter sp. and Pseudomonas aeruginosa respectively (fig. 1).
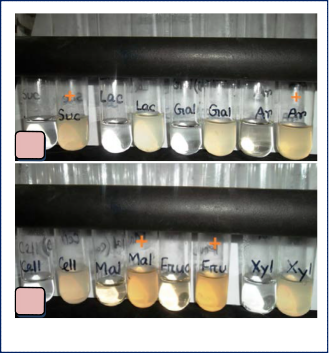
Morphological observation indicated the bacteria were found to be gram positive coccus, aerobic, non-motile and viable cell count was 459×107 CFU/ml (fig. 2). Isolated bacterium was lactose non-fermenting (fig. 3) and was unable to degrade cellulose but had the ability of utilizing arabinose, sucrose, maltose, fructose as carbon sources (fig. 4a-b). Bacteria had citrate metabolism ability (fig. 5), catalase activity (fig. 6) and also the ability of breaking
down the amino acid tryptophan into indole (table. 1). The bacterium was multi-drug resistant (table. 2) and found to be resistant to amoxicillin, penicillin and vancomycin (fig. 7a-b). Minimum Inhibitory Concentration (MIC) value against vancomycin was 50µg/ml (fig. 8).
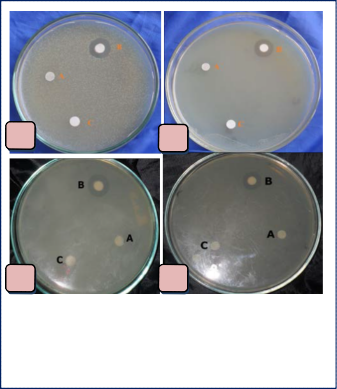


Fig. 1. (a) Zone of inhibition against Agrobacterium sp. (b) Zone of inhibition against E.coli BL21 (c) Zone of inhibition against Enterobacter sp. (d) Zone of inhibition against Pseudomonas aeruginosa.

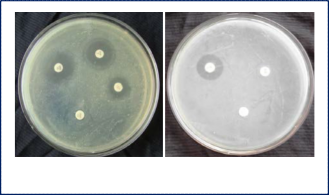


Fig. 8. Minimum inhibitory concentration (C2 = 50µg/ml).
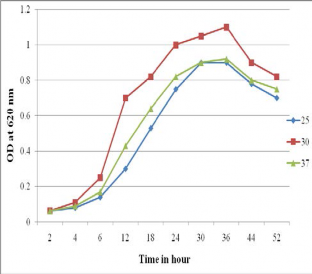
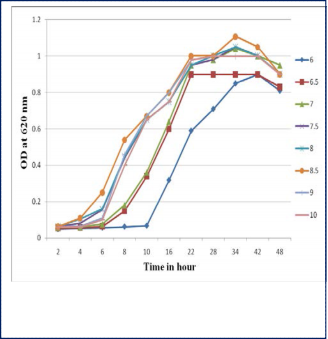
The bacterial growth depends on temperature and pH. Total eight different pH parameters were used to examine bacterial growth. Growing ability of the bacteria was observed from pH 6 to pH 10 while culturing on different pH (fig. 9). The optimum pH for the growth of the isolates was 8.5 and the temperature was 300C (fig. 10).
Biochemical Tests
Tests | Results |
Gram | + |
Motility | - |
Methyl Red (MR) | + |
Catalase test | + |
SIM test: Sulfide Indole Motility | - + - |
Simmons citrate agar test | + |
Lactose fermentation test | - |
Galactose | - |
Arabinose | + |
Sucrose | + |
Lactose | - |
Maltose | + |
Fructose | + |
Xylose | - |

Antibiotic sensitivity test
Antibiotics | Range of antibiotics | R | S and I |
Amoxicillin | 2mm | R | - |
Penicillin | 0mm | R | - |
Erythromycin | 12mm | - | I |
Vancomycin | 0mm | R | - |
Gentamycin | 20mm | - | S |
Ciprofloxacin | 17mm | - | S |
chloramphenicol | 18mm | - | S |
(5~10mm) = Resistance to antibiotic (R), (10+~15mm) = Intermediate resistance (I), (15+~20mm) = Sensitive to antibiotic (S).
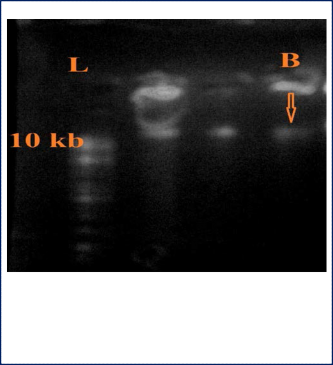
After extraction of the DNA it is essential to check its quality and integrity. Usual laboratory practice of checking DNA purity is to run the extracted DNA on a 1% agarose gel. Molecular weight of isolated bacterial genomic DNA showing a band above 10 kb size with not much of smearing indicated good integrity of DNA (fig. 11).
Antibiosis is considered as the most common mechanisms of action deployed by microorganisms up to now investigated [20]. Antibiosis is defined as antagonism brought about the metabolites of fungal antibiotics or antibiotic like compounds, lytic enzymes, volatile compounds or other toxic substances. It is relatively easy to demonstrate inhibition of one microorganism by another on agar and evidence of strain dependence in the effect [21]. The isolate was screened out as antagonistic bacterium though this physical parameter. Antagonistic test of bacteria was performed against Agrobacterium tumefaciens, E.coli BL21, Enterobacter sp. and Pseudomonas aeruginosa where 20µl concentration of the isolated bacterium used against each of the four test organisms.
Actually, most of the early studies on biological control of plant pathogens, being based on in vitro screening of potential biocontrol candidates, tended to select for microorganisms able to produce toxic metabolites, since the appearance of an inhibition zone was considered as factor for screening [22]. In the present study, the isolate was observed with 14mm, 14mm, 10mm and 13mm zone of inhibition against Agrobacterium tumefaciens, E.coli BL21, Enterobacter sp. and Pseudomonas aeruginosa respectively.
The antagonistic activities of L. fermentum, L. plantarum and L. casei were found against P. aeruginosa and E. coli. In 2011, Shanthya reported that 10mm, 13mm and 10mm of inhibition zones were found against three E. coli strains and 9mm, 18mm of inhibition zones were found against P. aeruginosa by L. fermentum and 11.5mm, 7mm, 9.5mm zone of inhibition against three E. coli strain and 6mm, 10mm zone against P. aeruginosa were formed by L. casei [23]. He also confirmed that 15.5mm, 20mm zone against two strains of P. aeruginosa and 12mm, 13mm, 11.5mm zone of inhibition against three strains E. coli were observed with
L. plantarum after 24 hours of incubation through disk
diffusion method. In 2003, Ya-Ping found the average antagonistic diameter of 14 mm against three strains of Escherchia coli [24]. Similarly, in this study 14mm zone of inhibition was found against E. coli BL21 strain by the isolate and 13mm zone of inhibition against P. aeruginosa and these results were significant with those of the references.
On the other hand, 10mm zone of inhibition against Enterobacter sp. was observed with the bacterium through disk diffusion method. Agrobacterium tumefaciens can be prevented by biocontrol organism A. radiobacter by soaking seeds or dipping transplants. A. tumefaciens is prevented due to the production of the antibiotic agrocin 84 by strain K84 of A. radiobacter [25, 26]. Similarly, A. tumefaciens had been inhibited by the isolate and the zone of inhibition was observed 14 mm indicating antagonistic effect.
The identification and the biological, molecular characterization of microorganisms, useful as biocontrol agents or as producers of bioactive compounds, are of great relevance for the modern and echo-compatible agriculture [27]. In the present research project, the morphological, physiological, biochemical and molecular characterizations were conducted for bacterial identification. The isolated bacterium was gram positive, staphylococci and utilized arabinose, sucrose, maltose, fructose as carbon sources. The bacteria were found to have citrate metabolism ability, breaking down ability of the amino acid tryptophan into indole and catalase activity. Band of bacterial genomic DNA above the 10 kb size with not much of smearing is the indication of good DNA integrity.
Globally, tones of antibiotics have been distributed in the biosphere during an antibiotic era of only about 60 years duration leading to resistant pathogens. With the development and spread of resistant bacteria and disease causing agents, importance of antagonistic bacteria is increasing simultaneously and applications of antagonistic bacteria are well documented. Bacteria with antagonistic property are screened out through this research work. Significant level of antagonistic effect was reflected through the bacteria. Such properties shown by the bacteria are considered as important factors in agriculture, aquaculture as well as biotechnology. The bacteria could be used as probiotics and alternative to antibiotics in future through further research and modifications.
The authors would like to express gratitude to Microbiology Laboratory, Department of Genetic Engineering and Biotechnology, University of Rajshahi, Rajshahi-6205, Bangladesh.
[1] T.L. Cza´ra´n, R.F. Hoekstra, and L. Pagie, Chemical warfare between microbes promotes biodiversity, Proc. Natl. Acad. Sci., vol.99, pp.786–790, 2002.
[2] B.C. Kirkup, and M.A. Riley, Antibiotic-mediated antagonism leads to a bacterial game of rock-paper – scissors in vivo, Nature, vol.428, pp.412–414, 2004.
[3] P. Jensen, and W. Fencial, Marine microorganisms and drug discovery current status and future potential. In Drugs from the sea, Fusetani N, Karger, Basel, Switzerland. 2000.
[4] P.R. Burkholder, L.M. Burkholder, and L.R.
Almodovar, Antibiotic activity of some marine algae of Puerto Rico, Bot. Mar., vol.2, pp.149-156, 1966.
[5] G.L. Patterson, and C.M. Bolis, Fungal cell wall polysaccharides elicit an antifungal secondary metabolite (Phytoalexin) in the cyanobacterium Scytonema ocellatum, J. Phycol., vol. 33, pp. 54-60, 1997.
[6] J.Trischman, D.M. Tapiolas, W. Fencial, R. Dwightand, and P.R, Jensen Salimanide A and B anti inflamatory depsipepdies from a marine Streptomycetes, J. Am. Chem. Soc. , vol.116, pp. 757-758. 1994.
[7] J.Gerard, P. Haden, M.T. Kelly, and A.J. Andersen,
Loloatin B- a cyclic decapeptide antibiotic produced in culture by a tropical marine bacterium, J. Nat. Prod., vol.62, pp.80-85 , 1996.
[8] N, Imamura, M, Nishijima, Adachi K. Takadera, M. Sakai, H. Sano, New anticancer antibiotics pelagiomicins producted by a new marine bacterium Pelagiobacter Variabillis, J. Antibiot., vol.50 pp.8-12, 1997.
[9] M. Slattery, I. Rajbhandari, and K. Wesson,
Competition mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis, Microb. Ecol., vol.41, pp. 90–96, 2001.
[10] J. Fabregas, A. Munoz, A. Otero, J.L. Barja, and M.
Roruaris, A preliminary study on antimicrobial
activities of some bacteria isolated from the marine environment, Nippon Suisan Gakkaishi, vol.57, pp.1377– 1380, 1991.
[11] P. Smith, M.P. Hiney, and O.B. Samuelsen, Bacterial
resistance to antimicrobial agents used in fish farming:
a critical evaluation of method and meaning, Ann. Rev. Fish Dis., vol. 4, pp.273–313, 1994.
[12]O.L. Akinbowale, H. Peng, M.D. Barton, Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia, J. Appl. Microbiol., vol. 100 pp. 1103–1113, 2006.
[13] R. Harikrishnan, C. Balasundaram, and M.S. Heo,
Lactobacillus sakei BK19 enriched diet enhances the
immunity status and disease resistance to streptococcosis infection in kelp grouper, Epinephelus bruneus, Fish Shellfish Immunol., vol. 29, pp.1037–1043, 2010.
[14] S.M. Aly, Y.A.G. Ahmed, A. Ghareeb, and M.F. Mohamed, Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of tilapia nilotica (Oreochromis niloticus) to challenge infections, Fish Shellfish Immunol, vol. 25, pp.128–136, 2008.
[15] R. Long, and F. Azam, Antagonistic interactions
among marine pelagic bacteria, Appl. Environ.
Microbial., vol. 67, pp. 4975–4983, 2001.
[16] J.G. Burgess, E.M. Jordan, M. Bregu, A. Mearns-
Spragg, K.G. Boyd Microbial antagonism: a neglected avenue of natural products research, J. Biotechnol., vol. 70, pp. 27–32. 1999.
[17] S. Emmanuel, J. Jebasingh, A. Murugan, Antagonistic activity of the barnacle (Balanus amphitrite) associated bacteria against human bacterial pathogens, World Journal of Medical Sciences, vol.6 no.1 pp.36-41, 2011.
[18] T. Islam, F. Sabrin, M.E. Islam, M.M. Billah, and
K.M.D. Islam, Analysis of antimicrobial activity of Lactobacillus paracasei sp. paracasei-1 isolated from regional yogurt, IRJALS., vol.1, no.4, pp.80-88, 2012.
[19] C. Barry, A. Ahmed and A.A. Khan, Endemic filiriasis
in Thakurgaon, East Pakistan. Amer. J. Trop. Med. Hyg.
, vol.22, no.4, pp.592-597,1976.
[20] C.R. Howell, The role of antibiosis in biocontrol,
Taylor and Francis, vol.2, pp.173-184,1998.
[21] T.M. Islam, Y. Hashidoko, A. Deora, T. Ito, and S.
Tahara, Suppression of dumping off disease in host
plants by rhizoplane bacterium Lysobacter sp. strain SB-K88 is linked to plant colonization and antibiosis against soilborne peronosporomycetes, Appl. Environ. Microbiol., vol.71, pp.3786-3796, 2005.
[22] R. Weissmann, and B. Gerhardson, Selective plant
growth suppression by shoot application of soil bacteria, Plant and Soil., vol. 234. pp.159-170, 2001
[23] R. Shanthya, S. Saranya, and N.H. Shenpagam, Antagonistic effects of Lactobacilli on gram negative bacteria, Journal of Advanced Laboratory Research in Biology, vol. 2, no.2, pp. 45-61, 2011.
[24] J. Ya-Ping, C. Wei-liang , Z. Rong , Z. Bai-rong, The
biological activity of antibacterial substance produced by Enterobacter cloacae B8, Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi., vol. 13, no.2 pp. 115-20,2003.
[25] G.N. Agrios, Plant Pathology, 3rd Ed. Academic Press Inc., London, pp.558-565, 1988.
[26] Collins, Soil borne plant pathogens, Department of Plant Pathology, North Carolina StateUniversity, pp728, 2001.
[27] D. Spadaro, and M.L. Gullino, Improving the efficacy
of biocontrol agents against soilborne pathogens, Crop Prot., vol. 24, pp.601-613, 2005.