International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1306
ISSN 2229-5518
Abstract-.
Globally, asymptomatic patients represent approximately 80% of total hepatitis C virus patients as indicated by Reverse-Transcription Polymerase Chain Reaction (RT-PCR) based viral genotyping. The present study was conducted to detect HCV genotypes and their prevalence among the asymptomatic populations from five Iraqi
governorates (Najaf, Babylon, Qadisyia, Karbala and Baghdad governorates), as the asymptomatic patients
globally represent about 80% from total HCV patients by using of viral genotyping through RT-PCR analysis. A875 samples were collected from different study groups. The total asymptomatic HCV positive cases by RT- PCR test were 103 out of 875, the genotype 4 was the predominant genotype that appeared in 89.4% of the pa- tients followed by genotypes 1b, 2b, 3a and 6a which had been found in 6.79%, 2.91%, 2.91% and 1.94% of the asymptomatic patients, respectively. The current study provide the first information in Iraq at least in the study areas about the presence of HCV genotypes 2b and 6a
Key words: asymptomatic,Genotype,HCV, investigation,prevalence,predominant,RT-PCR
IJ————S—————— E———————R———
Hepatitis C virus infection is one of the major public health problem in both developed and developing countries since discovering at
1989 (1,2). It is estimated that HCV infect 200 million peoples (3%) of the world’s population and there are at least 21.3 million HCV carriers in the Middle East and Eastern Mediterranean countries (3). The infection is more often asymptomatic. About 85%-90% of acute cases and 70%-80% of chronic cases are asymptomatic and the jaundice develops only in one third of the symptomatic patients (4,5).
The virus exhibit substantial genetic variation so there are at least eleven distinct viral genotypes : 1 through 11 , and variable number of sub- types found usually in specific geographical regions and display significant difference in their aggressiveness and in response to antiviral therapy (6,7). Genotypes 1, 2 and 3 are widely distributed throughout USA, Europe, Australia and East Asia; genotype 4 is largely confined to the Middle East, Egypt and central Africa; genotype 5 and 6 are found predominantly in South Africa
————————————————
• Author name *Ghanim A. Al-Mola: *Department of Biology College of
Science for Women, Babylon University, Babylon-Iraq
• E.Mil: almolaghanim@yahoo.com (corresponding author
Co-Author name **Hashim R. Tarish: _ raheem2004@yahoo.com
**KararM.Abdusada: kmabdulsada@ yahoo.com** Department of Microbiology College
and South East Asia, respectively (6,7,8); HCV genotypes 7, 8, and 9 have been identified mainly in Vietnamese patients; while genotypes
10 and 11 have been reported from Indonesia (9).
There are an increasing evidences that HCV genotypes possess different biological potentials, certain genotypes are more frequently associated with severe forms of liver disease and more amenable to interferon treatment make the genotyping of infecting virus is one of the prime predictors of the disease progression and the response to antiviral therapy. Consequently, typing of HCV isolates becomes an additional tool in diagnosis of the infection (10,11,12).
The distribution of HCV types in our country is still unclear because of the marked paucity in the studies about them, the purpose of this study was to estimate the prevalence of HCV genotypes among different asymptomatic study groups.
Methods:
Sampling
Serum samples were collected from a total of 875 clinically asymptomatic individuals were included in this study that
of Medicine-University of Kufa, Al-Najaf-Iraq
IJSER © 2013 http://www.ijser.org
*RafahHadyLateef: Hadiraf ah@yahoo.com
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1307
ISSN 2229-5518
performed from the beginning of June 2009 to the end of August
2010, 517 ofthese samples were randomly taken from asymptomatic individuals (general population) of all ages, both sexes, different residences and the occupations whom attended to hospitals from five Iraqi governorates; Najaf, Babylon, Qadisyia, Karbala and Baghdad governorates, 145 of samples were from thalassemic patients, 100 haemodialytic patients, 38 from the medical staff, 69 of blood donors (blood bank) whom positive for ELISA-II test and 6 were hepatocellular carcinoma patients from them also liver biopsies have taken. All samples were subjected to investigation of HCV genotypes by use of reverse transcription PCR (RT-PCR).
Asymptomatic HCV genotyping:
Genotyping through RT-PCR protocol consists of 4 main procedures: Extraction, Reverse transcription, Amplification and Detection.
1-Viral RNA extraction step have been done inside the biological cabinet
a- RNA Extraction from serum samples:
3- Amplification:Mixture sets of primers were used according to Ohnoet al. ( 13), the primer solutions were prepared by dissolving the lyophilized primers, Alpha DNA company, Canada in Tris-EDTA buffer as the instructions of the manufacturer. Two rounds of amplification had implemented, the products of first round were used in the second round .
4- Detection: Reaction products were analyzed by 2% agarose gel electrophoresis, 8 µl of amplification products added in each well. The products of the two mixture of primer were separately loaded. The DNA bands were observed through viewing under UV transilluminater, and pictures took by Bio-Document Analyzer,(Biometra, Turkey).
Results:
The total asymptomatic HCV positive cases by RT-PCR test were
103 out of 875, The genotyping by use of RT-PCR revealed that 89 patients were infected with genotype 4 from total 103 HCV positive
IJSER
It was implemented by use ofRibo Virus Columns Extraction
kit,Sacace Biotechnologies, Italy, the procedure was done according to the manufacturer’s instructions; at which 150 µl of serum was loaded to each Column followed by consecutive steps of washing
and finally the RNA that attached to the silica membrane in the Ribo Viruscolumns were dissolved in nuclease-free water and aggregated at button of Eppindorff tubes by centrifugation than plugged rapidly and stored at -70°C until used .
b- The extraction from liver biopsies was done through the Easy Express Viral Nucleic Acid Release Kit,( BiochainCompany,USA), the procedure was done according to the manufacturer’s instruction ;
100µl of thawed proteases solution was put in a sterile 0.5ml Eppindorff tubes, one mg of liver biopsies were added to each tube and mixed by vortex for 15 seconds followed by multiple rounds of heating and centrifugation, then the supernatant were aspirated and stored at -70 ºC until use.
2- Reverse Transcription: This process was done by used of Reverse Transcription System Kit,(Promega Corporation, USA). The procedure have been done inside the biological cabinet and the steps conducted according to manufacturer’s instructions, The frozen RNA samples directly thawed inside incubator at 70˚c for 10 minutes then added to a mixture from kit components, mixed and underwent several steps of heating cycles. The mixture hence contain cDNA ,
stored at -20˚C until use.
cases with percentages of 86.4% which indicates that genotype 4 is
the predominant type.
Genotype 1b rank below genotype 4 and appeared in 7 cases (6.79%), followed by both genotypes 2b and 3a, each appeared in 3 cases and percentage of 2.91%,where as genotype 6a was found only in two cases and showed the lowest rate among others (1.94%); One thalassemic patient has experienced a mixed infection with
HCV genotypes 4 and 1b. (Table-1), (Figure-1).
Discussion:
The new researches explained that the rate of development of persistent infection at the asymptomatic acute patients significantly higher than that of symptomatic patients, besides to about 80% of HCV patients were asymptomatic (14,15) ; therefore, it is important to know the present information of asymptomatic HCV infected subjects at our community. The variation in the geographical distribution among the viral genotypes in the world made it worthy to get a local information pertain viral genotypes as that considered as an epidemiological marker particularly in tracing the source of infection and elucidating the possible mode of transmission, assessment the duration and benefit of antiviral therapy and future development of vaccine (16).
In the our study we used a genotyping method through RT- PCRanalysis that has been originally employed by Ohnoet al. ( 13)
which involved wide range of HCV genotypes: 1a, 1b, 2a, 2b, 3a,
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1308
ISSN 2229-5518
3b, 4, 5a, and 6a. The current study was revealed that the genotype 4 is the predominant genotype which found in 86.4% of the infected cases followed by Genotypes 1b, 2b, 3a and 6a that came in percentages of 6.79%, 2.91%,2.91% and 1.94% respectively. This study was the first at least in the study areas that involved the searching of HCV genotypes among asymptomatic patients by using of RT-PCR test and the first that recorded presence of genotypes 2b and 6a in Iraq.
Al-Kubaisyet al.(17) have studied the sero-prevalence of HCV genotypes by use of genotype specific ELISA-III test searching antibodies at thalassemic Iraqi children, they were reported that genotype 4 was the most frequent type that has been found it in
35.4% of the infected cases followed by 1a in 27.1% of cases and genotype 1b that occurred in 22.9% of cases whereas mixed infection with genotypes 1a and 4 was set in 14.6% of the HCV among infected thalassemic children. Other study, by Al-Kubaisyet
al.(18) who used the same technique (genotype specific ELISA-III
genotype 1a was the most prevalent type which found in 40% of the patients whereas genotypes 1b and 4 were found in 33.3% and
26.6% of the HCV patients, respectively.
In Iran (non-Arabic neighboring country), the genotypic pattern was differed, for example: Zarkesh-Esfahaniet al. (23) had observed that the frequency of HCV genotypes was determined as follows: genotype 3a (61.2%), genotype 1a (29.5%), genotype 1b (5.1%), genotype 2 (2%) and mixed genotypes of 1a plus 3a (2%).
The difference in the frequency of HCV genotype from that in Jordan and Iran was in congruence with observations of Ramia and Eid-Fares (24) whom referred that the genotype 4 was the prevalent type at the Arabic Middle Eastern countries (except for Jordan) and genotype 1 was dominant at non-Arabic Middle East countries.
In Turkey (non-Arabic neighboring country), the genotype 1 was the predominant genotype which has been observed in 92% of the
patients, followed by genotype 4 in 5% and genotype 2 in 2% of the
IJSER
test searching antibodies) and observed differed result among
haemophiliac patients in Baghdad and co-infected with the human immunodeficiency virus (HIV), they referred that the genotype 1a was the dominant genotype followed by genotypes 1b and 4.
The above difference in genotype patterns in Iraq may be attributed to the test that has been used in the diagnosis (ELISA test searching antibodies), number of samples, type of the patients (haemphiliac and thalassemic and not asymptomatic) and the co- infection with HIV.
In Saudi Arabia (Arabic neighboring country), similar finding was showed byShobokshi et al. (19) who found that the majority (62%) of the HCV infections in the Saudis were with genotype 4, while
the other genotypes found with a lesser extent as follows: 1 (24.1%); 2 (7.4%); 3 (5.9%); and 5 (0.3%). In Syria, the frequency of HCV genotypes was similar to that of the current study, Antakiet al. (20) had pointed out that genotype 4 was the most prevalent type at the Syrian HCV patients it was present in 59% of the patients followed by genotype 1 ( in 28.5% of patients) and genotype 5 that found in 10% of the infected cases.
In Kuwait, Pasacaet al. (21) also observed similar result, they were recorded that genotype 4 was the predominant genotype followed by genotype 1. Where as In Jordan, Bdour(22) had drew absolutely varied picture from that in above Arabic counties, she studied HCV
genotypes at the Jordanian haemodialysis patients and found that
patients (25).
The interpretation of the presence of genotypes 1b, 2b and 3a in some Iraqi patients in the current study is that these types might had been introduced to our country from Iran, Jordan and Turkey where the prevalence of these genotypes was relatively high as a result of traveling, immigration and the matrimonies.
Genotype 4 was found to be the predominant genotype among the HCV-infected Lebanese patients (53.3%), genotype 1a was ranking below in 43.3% of infected cases and genotype 1b found in 8% of the patients (26). This result was in agreement with our study result concerning the dominancy of genotype 4 amongst other types at HCV patients. Similar finding was also reported by Ohnoet al. (27) at the HCV Yemeni patients where genotype 4 was the most prevalent type followed by genotype 1.
In Egypt, similar result was obtained by El-Kady et al. (28), they observed that the genotype 4 was detected in 89.6% followed by genotype 1 in 7.7% of the patients. The prevalence of genotype 4 in the Egyptian HCV patients was quite similar to that of our study and the majority of other Arabic middle Eastern countries too, that may attributed to similarity in the peoples traditions, their ethnicity and
the geographical vicinity. In Libya, which is an Arabic North-African country, the genotype
4 was the commonest and appeared in 35.7% of the HCV patients,
genotype 1 was came in a lesser frequency (32.6%) followed by
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1309
ISSN 2229-5518
genotypes 2 and 3 which has been found at 16.7% and 13.2% of the patients, respectively ( 29). The dominancy of genotype 4 was not only in consistence with the our result but also with Egypt and most other middle-Eastern Arabic countries that may attributed to the traveling, working and the mating.
However the pattern of genotypes' frequencies at the other three Arabic north African counties ( Tunisia, Morocco and Algeria) was extremely differed; the genotype 1 and 2 were the dominant genotypes while genotype 4 has a trivial frequencies (6,30,31). Idresset al. (32) had reported that the genotype 3a was the dominant genotype (34.1%) at the Pakistani HCV patients followed by 2a (8.1%) , 3b (7%) and 1a (5.4%). That similar to the result of
Vermaet al. (33) in India who were reported that genotype 3 is the prevalent type (40.81%) while the other genotypes; 1a, 1b and mixed of 1 + 3 have been found in prevalence of 6.12%, 8.16% and 17.3% in HCV patients, respectively.
The presence of genotype 3a (which is the prevalent type in the
%). In Brazil which is a south American country, Sawada et al.
(39) had registered that genotype 1 was the predominant type which came in percentage 94% of HCV patients while genotype 3 has
been recorded at 6% of the patients.
It is of interest to note that the recording of genotypes 1b and 2b in our study at frequency of 2.91% for each of them may be attributed to the transmission of these genotypes from other regions such as European or American countries where the prevalence of these types was high, might as a result of immigration, traveling, outside treated patients, importing of contaminated blood or the blood products, this finding was in congruence with the observations of Sy and Jamal
(3) who were mentioned that these genotypes have a cosmopolitan distribution but the highest frequencies were at Europe and America.
Conclusions
In the present study, we concluded that the HCV genotype four was
Indian subcontinent) in our studIy at aboJut 2.91% of HSCV Iraqi ER
patients may be that this genotype has been introduced to our country by traveling.(33)
In Indonesia which is one of southeast Asian countries, Utamaet
al. (34) were registered that genotype 1b was the most frequent type which was found in 47.3% of HCV patients. Genotype 1 also prevalent in Japan Whereas the viral genotypes 6, 7, 8, 9, 10 and 11 were observed in Vietnam, Thailand, Myanmar and Philippines (6,24).
In the Gabon which is located at the middle of Africa , Ndong- Atome et al. (35) were reported that genotype 4 was the most dominant type that had been recorded in 90.9% of HCV patients, this frequency was quite similar to that of our study whilst in South Africa, Vardas et al. (36) had registered that genotype 5 was dominant followed by genotype 1a.
In the current study the genotype 5 was not observed at any of the HCV patients, that may be due to the limitation in the geographical distribution of this genotype which was found mainly at the south African countries(24).
In Spain, Torres-Puente et al. (37) had recorded that genotype 1 was the predominant followed by types 2 and 3. In United States, Eysteret al. (38) had showed that the genotype 3a was prominent type which found in 41% of the patients followed by genotypes 1a
and 1b that appeared in 31% and 13% of the patients, respectively.
the predominant genotype among asymptomatic HCV patients in the
study areas( Najaf,Babylon,Qadisyia,Karbala and Baghdad Iraqi governorates). This study also recorded that the HCV genotypes 2b and 6a circulating among asymptomatic patients in Iraq.
Author’s contributions
Ghanim A.A. and HashimR.T.conceived the study. Karar M.A and Ghanim A.A. collected the samples and perform the molecular analysis. Hashim R.T. and KararM.A.andRafah H.L. search literature and drafted the manuscript. Ghanim A.A. reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Thanks to all the clinicians and patients for their cooperation in the study.
References:
1-Choo,Q.L; Weiner,A.J; Overby,L.R; Kuo,G; Houghton,M. and
Bradley,D.W. (1989). Hepatitis C virus: the major causative agent of viral non-A, non-B hepatitis. Br.Med.Bull. 46:423-
41.
2-Greenwood,D; Slack ,R; Peuther, J.B. and Barer, M. (2007). Medical Microbiology.17th Edit. Elsevier Churchill
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1310
ISSN 2229-5518
Livingstone. Pp.
3-Sy,T. and Jamal, M. M. (2006). Review: Epidemiology of
Hepatitis C Virus (HCV) Infection.Int.J.Med.Sci. 3(2):41-46.
4-Bruno,S. and Facciotto,C. (2008 ). Review: The natural course of
HCV infection and the need for treatment. Ann.Hepatol.
7(2):114-19.
5-Miyazaki,T; Honda,A; Ikegami,T; Hara,T; Saitoh,Y; Hirayama,T; Doy,M. and Matsuzaki,Y. (2009). The associated markers and their limitations for the primary screening of HCV carriers in public health examination.Hepatol.Res. 39(7):664-74.
6-Simmonds,P; Bukh,J; Combet,C; Deleage,G; Enomoto,N; Feinstone,H; Halfon,P; Inchauspe,G; Kuiken,C; Maertens,G; Mizokami,M; Murphy,D.G; Okamoto,H; Pawlotsky,J.M; Penin,F; Sablon,E; Shin-I,T; Stuyver,L.J; Thiel,H.J; Viazov,S; Weiner,A.J. and Widell,A. (2005).
Consensus proposals for a unified system of nomenclature of
Evidence of recombination in quasispecies population of a Hepatitis C virus patient undergoing anti-viral therapy.Virol.J. 3:87-93.
13-Ohno,T; Mizokami,M; Wu,R.R; Salih,M.G; Ohba,K.I; Orito,E; Mukaide,M; Williams,R;. andLau,J.Y.(1997). New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4,
5a, and 6a. J.Clin.Microbiol. 35(1):201-7.
14-Contreras,A.M; Ochoa-Jimenez,R.J; Celis,A; Mendez,C; Olivares,L; Rebolled,C.E; Hernandez-Lugo,I; Aguirre- Zavala,A.L; Jemenez-Mendez,R. and Chung,R.T. (2010). High antibody level: an accurate serological marker of
viremia in asymptomatic people with hepatitis C infection. Transfusion. 50(6):1335-43.
15-Grammatikos,G. and Sarrazin,C. (2010). Review: Chronic
hepatitis C. Dtsch.Med.Wochenschr. 135(50):2525-34.
IJSER
hepatitis C virus genotypes.Hepatology. 42:962-73.
7-Argentini,C; Genovese,D; Dettori,S. and Rapicettta,M. (2009). Review: HCV genetic variability: from quasispecies
evolution to genotype classification. Futur.Microbiol. 4:359-
73.
8-Agha,S; Tanaka,Y; Sandy,N; Kurbanov,F; Abo-Zeid,M; El- Makey; Khalaf,M; Ohta,N; Yoshizawa,H. and
Mizokami,M. (2004). Reliability of hepatitis C virus core
antigen assay for detection of viremia in HCV genotypes 1, 2,
3 and 4 infected blood donors; collaborative study between
Japan Egypt and Uzbekistan. J.Med.Virol. 73:216-222.
9-Lee,C.M; Hung,C.H; Lu,S,N. and Changchien,C.S. (2008). Review: Hepatitis C virus genotypes: clinical relevance and therapeutic implications. Chan.Gun.Med.J. 31(1):16-25.
10-Hadziyannis,S.J; Sette,H; Morgan,T.R;Casane,D. and Lu,S,N. (2004). Peginterferon- alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann.Intern.Med. 140: 346-355.
11-Bowden,D.S. and Berzsenyi,M.D. (2006). Review: Chronic hepatitis C virus infections : genotyping and its clinical role. Futur.Microbiol. 1:103-12.
12-Moreno,M.P; Casane,D; Lopez,L. and Cristina,J. (2006).
16-Jacobson,I.M; Davis,G.L; El-Serag,H; Negro,F. and Trepo,C.
(2010). Prevalence and challenges of liver diseases in patients with chronic hepatitis C virus infection.Clin.Gastroenterol.Hepatol. 8(11):924-33.
17-Al-Kubaisy,W.A; Al-Naib,K.T. and Habib,M.A. (2006b). Prevalence of HCV/HIV co-infection among hemophilia patients in Baghdad.Eas.Mediterr.Heal.J. 12(3-4):264-69.
18-Al-Kubaisy,W.A; Niazi,A.D. and Kubba,K. (2002). History of miscarriage as a risk factor for hepatitis C infection in
pregnant Iraqi women.Eas.Mediterr.Heal.J. 8(2-3):239-44.
19-Shobokshi,O.A; Serebour,F.E. and Skakni,L.I.(2003). Hepatitis C virusgenotypes/subtypes among chronic hepatitis patients in Saudi Arabia.Saudi.Med.J. 24(2):87-91.
20-Antaki,N; Haddad,M; Kebbewar,K; Abdelwahab,J; Hamed,O; Araj,R; Alhai,N; Haffar,S; Assil,M; Ftayeh,M; Assaad,F; Doghman,D; Ali,T; Nasserelddine,M. and Antaki,F. (2009). The unexpected discovery of a focus of hepatitis C virus genotype 5 in a Syrian province.Epidemiol.Infect. 137(1):79-84.
21-Pasaca,A; Al-Mufti,S; Chugh,T and Said-Adi,G.(2001). Genotypes of hepatitis C virus in Kuwait.Med.Princ.Pract.
10:55-57.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1311
ISSN 2229-5518
22-Bdour,S. (2002). Hepatitis C virus infection in Jordanian haemodialysis units: serological diagnosis and genotyping. J.Med.Microbiol. 51:700-04.
23-Zarkesh-Esfahani,S.H; Kardi,M.T. and Edalati,M. (2010). Hepatitis C virus genotypes frequency in Isfahan province of Iran: A descriptive cross-sectional study. Virol.J. 7:69-75.
24-Ramia,S. andEid-Fares,J. (2006). Review: Distribution of hepatitis C virus genotypes in the Middle East. Int.Infec.Dis.10(4):272-77.
25-Sanlidag,T; Akcali,S; Ozbakkaloglu,B; Ertekin,D. and Akduman,E. (2009). Distribution of hepatitis C virus genotypes in Manisaregion,Turkey. Mikrobiyol.Bul.
43(4):613-18[Abstract].
26-Ramia,S; Ramlawi,F; Eid-Fares,J; Klayme,S. and Naman,R.
(2007). Genotypes of hepatitis C virus (HCV) among positive
f hepatitis C in the Middle East. Saudi.J.Kidne.Dis.Transplant.
22(1):1-9.32-Idress,I.M; Ahmed,H; Ghafoor,S; Ali,M; Ali,L. and Ahmed,A. (2011). Hepatitis C virus genotypes circulating in distinct Swat of Khyber Pakhtoonkhaw, Pakistan.Virol.J. 8(16):16-21.
33-Verma,V; Chakravarti,A. and Kar,P. (2008). Genotypic characterization of hepatitis C virus and its significance in patients with chronic liver disease from Northern India.Diagn.Microbiol.Infect.Dis. 61(4):408-14.
34-Utama,A; Tania,N.P; Dhenni,R; Gani,R.A; Hasan,I; Sanityoso,A; Lelosutan,S.A; Martamala,R; Lesmana,L.A; Sulaiman,A. and Tai,S. (2010). Genotype diversity of hepatitis C virus (HCV)-associated liver disease patients in Indonesia. Liv.Int. 30(8):1152-60. 35-Ndong-Atome,G.R; Makuwa,M; Ouwe-Boyer,O; Pybus,O.G; Branger,M; LeHello,S; Boye-Cheik,S.B; Brun-Vezinet,M; Kazanji,M; Roques,P. and Bisser,S.(2008). High prevalence of hepatitis
C virus infection and predominance of genotype 4 in rural
IJSER
Lebanese patients: comparison of data with that from other
Middle Eastern countries. Epidemiol.Infect. 135:427-32.
27-Ohno,T; Mizokami,M; Saleh,M.G; Orito,E; Ohba,K.I; Wu,R.R; Koide,T; Tibbs,C.J; Nouri-Aria,K.T; Tokudoms,S. and Williams,R. (1996). Usefulness and limitation of phylogenetic analysis for hepatitis C virus core region: application to isolates from Egyptian and Yemeni patients. Arch.Virol. 141(6):1101-13.
28-El-Kady,A; Tanaka,Y; Kurbanov,F; Sugauchi,F; Sugiyama,M; Khan,A; Sayed,D; Moustafa,G; Abdel- Hameed,A.R. and Mizokami,M. (2009). Genetic variability of hepatitis C virus in South Egypt and its possible clinical implication.J.Med.Virol. 81(6):1015-23.
29-Elasifer,H.A; Agnnyia,Y.M; Al-Alagi,B.A. and Daw,M. (2010). Epidemiological manifestations of hepatitis C virus
genotypes and its association with potential risk factors
among Libyan patients.Virol.J. 7:317-24.
30-Hmaied,F; Ben-Mamou,M; Saune-Sandres,K; Rostaing,L; Slim,A; Arrouji,Z; Ben-Redjeb,S. and Izopet,J. (2006). Hepatitis C virus infection among dialysis patients in Tunisia: incidence and molecular evidence for nosocomial
transmission. J.Med.Virol. 78(2):185-91.
31-Fallahian,F. and Najafi,A. (2011). Review: Epidemiology o
Gabon.J.Med.Virol. 80(9):1581-87.
36-Vardas,E; Sitas,F; Seidel,K; Casteling,A. and Sim,J. (1999). Prevalence of hepatitis C virus antibodies and genotypes in asymptomatic, first-time blood donors in Namibia. Bull.Worl.Health.Org. 77(12):965-72. 37-Torres-Puente,M; Cuevas,J.M; Jimenez-Harnandez,N; Bracho,M.A;
Garcia-Robles,I; Wrobel,B; Carnicer,F; Del-Olmo,J; Ortega,E; Moya,A. and Gonzalez-Candelas,F. (2008). Genetic variability in hepatitis C virus and its role in antiviral treatment response.J.Viral.Hepatol. 15(3):188-99.
38-Eyster,M.E; Sherman,K.E; Goedert,J.J; Katsoulidou,A. and
Hatzakis,A. (1999). Prevalence and changes in hepatitis C virus genotypes among multi-transfused persons with hemophilia.The Multicenter Hemophilia Cohort Study.J.Infect.Dis. 179(5):1062-69.
39-Sawada,L; Pinheiro,A.C; Locks,D; Pimenta,S; Rezende,P.R; Crespo,D.M; Crescente,J.A; Lemos,J.A. and Oliveira,F.A. (2011).Distribution of hepatitis C virus genotypes among different exposure categories in the State of Pará, Brazilian Amazon.Res.Soc.Bras.Med.Trop. 44(1):8-12
.
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1312
ISSN 2229-5518

Mixture 1
DNA Ladder 338 bp 338 bp 234 bp 234 bp 234 bp
Mixture 2
IJSER
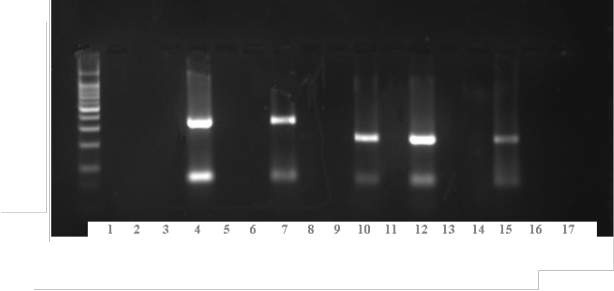
Transcription Polymerase Chain Reaction by Using of Two Mix- turesof Primers
Mixture 1: Lanes 4 and 7 genotype 2b
Lanes 10, 12, and 15 genotype 1b
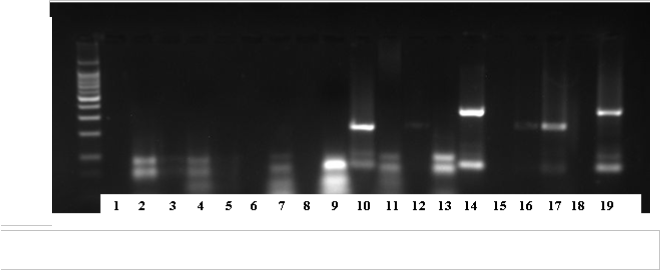
Mixture 2: Lane 9 genotype 4
Lanes 10 and 17 genotype 3a
Lanes 14 and 19 genotype 6a
IJSER © 2013 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 4, Issue 12, December-2013 1313
ISSN 2229-5518
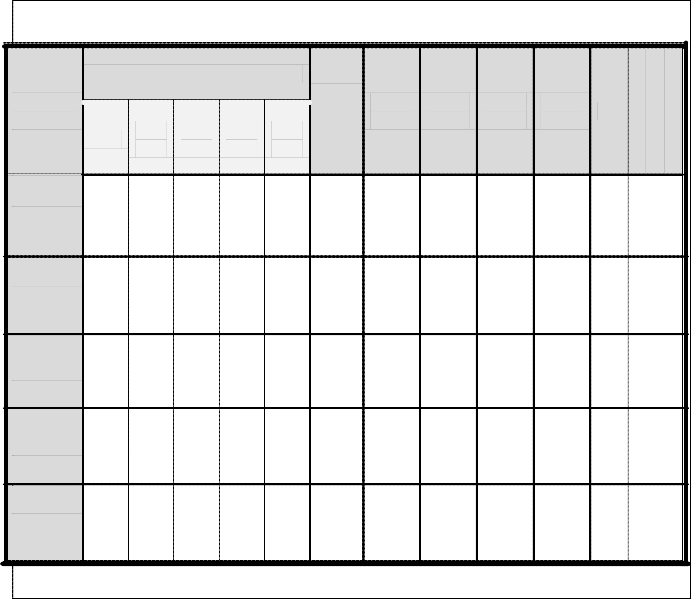
Table (1): The Results of genotyping of asymptomatic HCV by Using RT-PCR Analysis in the Study Groups
Genotypes
(G)
Normal asymptomatic indviduals
Thalas- se-mic
Medical staff
HCC. patients
Renal dialysis
Blood donors
Total
Najaf
Baby-
lon
Qa-
disyia
Kar-
bala
Bagh-
dad
patients
G.1b 1 1 0 0 0 2 0 0 0 3 7 6.79%
G.2b 0 0 0 0 0 2 0 0 0 1 3 2.91%
G.3a 1 0
I0 J0
S1 1
E0 0
R0 0 3 2.91%
G.4 5 3 2 3 1 21* 2 4 7 41 89 86.4%
G.6a 0 0 0 0 0 2 0 0 0 0 2 1.94%
* Genotypes mixed infection (G.1b + G.4).
IJSER © 2013 http://www.ijser.org