
International Journal of Scientific & Engineering Research, Volume 6, Issue 1, January-2015 1301
ISSN 2229-5518
Identifying the ARR3 gene responsible for a cone dystrophy using genetic browsers
Hayder Muhsin Khalfa , Ameer Ali Shakir , Noor Hassan Khalfa
Abstract—This study aims to identify the gene responsible for a cone dystrophy within a family. Prioritizing several possible genes that can be associated with cone dystrophies this task investigates the gene with cone associated function and highest expression in the retina. ARR3 gene has been found to have the highest expression within the retina compared to other genes found on the locus of the X chromosome. Using different website and electronic genetic databases the section on the x chromosome has to be identified and examined. Identification of the genes in the region on the X chromosome is essential since all genes have to be analyzed. Each gene found need to be assessed in terms of function and region of expression within the human body. Genes expressed in the retina posing a photoreceptor function are prioritized to narrow down the focus on a particular gene. Top priortised genes are also assessed on how many EST (expressed sequence tags) they have. Top priortised genes are also assessed on how many EST (expressed sequence tags) they have. After the top gene has been selected thorough analysis is required to identify the function of the gene, this is achieved using literature search to determine the association of the gene and photoreceptors. The selected gene contains exons which are the coding regions of the gene, primers have to be designed to amplify the sequence of each exon present in the gene. ARR3-201 transcript of ARR3 gene found in the region between the two different microsatellite markers provided has been analyzed and primers have been designed to amplify its exons. Evidence has been provided in the literature that this gene is localized in cone photoreceptors and its associated with different functions in the cones photoreceptors furthermore the expression rate was significantly high in the retina which provides more evidence that ARR3 gene is possibly associated with cone dystrophy identified within the family.
Index Terms— ARR3, Cone Dystrophy, Gene, X chromosome, Genetic Browsers, Sequence Tags, EST.
—————————— ——————————
his study aims to identify the gene responsible for a cone dystrophy within a family. Prioritizing several possible
genes that can be associated with cone dystrophies this task investigates the gene with cone associated function and highest expression in the retina. ARR3 gene has been found to have the highest expression within the retina compared to other genes found on the locus of the X chromosome. To aid the process of genetic identification of possible mutations that occurred within the ARR3 gene a pair of primers have been designed for each exon present in the ARR3 gene.
This task follows a particular strategy to identify the gene re- sponsible for the cone dystrophy. As suggested by the infor- mation provided the cone dystrophy is X linked and responsi- ble genes fall in between two possible microsatellite markers. Using different website and electronic genetic databases the section on the x chromosome has to be identified and exam- ined. Identification of the genes in the region on the X chro- mosome is essential since all genes have to be analyzed. Each gene found need to be assessed in terms of function and re- gion of expression within the human body. Genes expressed in the retina posing a photoreceptor function are prioritized to narrow down the focus on a particular gene. Top priortised genes are also assessed on how many EST (expressed se- quence tags) they have. After the top gene has been selected thorough analysis is required to identify the function of the gene, this is achieved using literature search to determine the association of the gene and photoreceptors. The selected gene contains exons which are the coding regions of the gene, pri- mers have to be designed to amplify the sequence of each exon present in the gene.
The region identified by the microsatellite markers provided and has been located on the X chromosome between
67,251,011-70,436,328 and segregating region Xq12-21. Be- low is the identified section on the X chromosome which con-
tains the suspected gene responsible for the cone dystrophy.
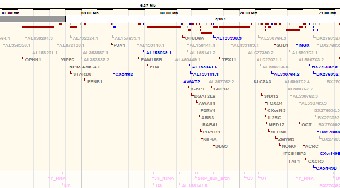
Below is a table showing the genes within the identified region on the X chromosome. The table contains the formal name of the gene and its expression in the human body. The expres- sion of each gene is associated with the prioritizing of the top genes responsible for the cone dystrophy in the family (3).
IJSER © 2015 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 6, Issue 1, January-2015 1302
ISSN 2229-5518
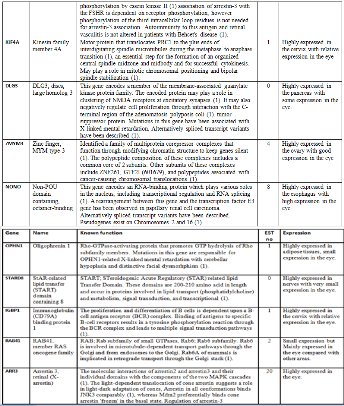
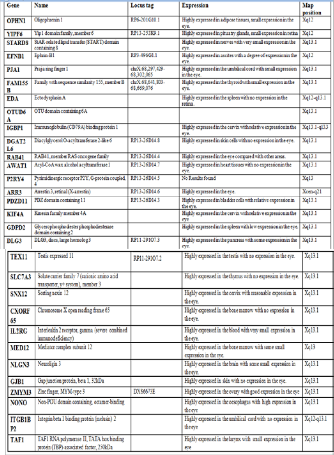
Below is a table showing the top nine genes that have been selected after a thorough analysis of their expression and func- tion using different genetic web browsers. Each gene in the table was investigated using the eye browse from the NEIBANK (3) browser to determine the number of EST (ex- pressed sequence tags) each gene had. The number of EST has been counted manually for each gene.
Below is a table showing the order of the prioritized top 9 genes and the known function of the gene as well as the pro- cesses involved in. The order of genes has been determined by the number of EST each gene has and the function it pre- formed in the eye. After an intense investigation I have found the above a meaningful prioritization since most of the genes selected have a high expression and high EST compared with other genes found on the section of the X chromosome.
————————————————
• Ameer Ali Shakir: Department of Biology, Faculty of science, Kufa univer- sity
• Noor Hassan Khalfa: Department of Computer Science, College of science for Girls, Babylon University, Iraq
IJSER © 2015 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 6, Issue 1, January-2015 1303
ISSN 2229-5518
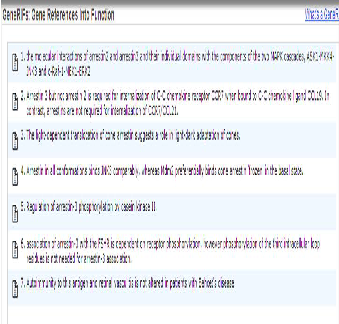
The gene selected as a top candidate gene is ARR3 gene and this is because the above mentioned function of the arrestin gene identify it to be a suitable candidate gene for the primer design furthermore its function is associated with sensory per- ception, signal transduction and visual perception which all are the characteristics of the cones photoreceptors. Below is a diagram showing the different transcripts of the ARR3 gene. The ARR3 gene has been selected as top candidate gene to investigate in order to determine the gene responsible for the cone dystrophy in the family. The selection of the ARR3 gene has been determined by the number of EST it has as well as the expression and functionality. The ARR3 gene has 20 EST and was highly expressed in the retina furthermore literature search exposed its association with cone photoreceptors. The ARR3 gene falls in the following region Chromosome X:
69,488,155-69,501,690(2).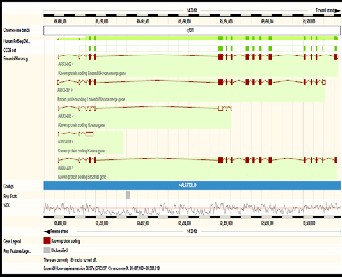
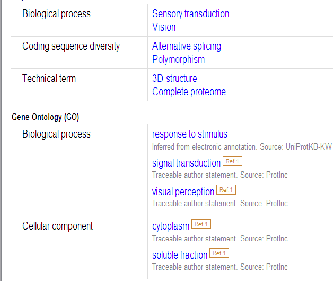
Below is a series of screenshots taken from the NCBI browser showing further details on the ARR3 gene. The ARR3 gene is associated with cone photoreceptors and its biological process is associated with visual perception, signal transduction and responses to a stimulus. There are several articles which pro- vide evidence to prove that the ARR3 gene is associated with the cones photoreceptors as shown in the above screenshot. Below is a screen shot showing the expression location of the arrestin gene and as shown it’s mainly and highly expressed in the eye.
IJSER © 2015 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 6, Issue 1, January-2015 1304
ISSN 2229-5518

contains 17 different exons which require a forward and backward primer in order to amplify.
Below is an image of the ARR3-201 transcript selected for analysis and primer design.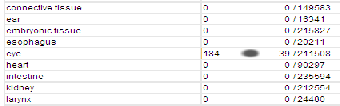
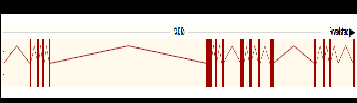
Below is a table showing the number of transcripts within the ARR3 gene. Highlighted in red is the transcript selected to be analyzed in order to design primers for each exon it contains. The ARR3-201 transcript have been selected since it contains the highest number of exons furthermore it’s a protein coding transcript and it covers most of the different exons found on other transcript in the ARR3 gene. The ARR3-201 transcript
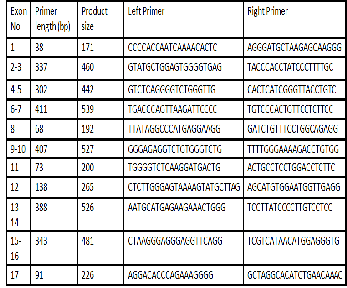
Below is table of left and right primers designed to amplify the different exons on the ARR3-201 transcript. The primers have been designed using the geneomic sequence and CDNA se- quence of ARR3-201 transcript.
IJSER © 2015 http://www.ijser.org
International Journal of Scientific & Engineering Research, Volume 6, Issue 1, January-2015 1305
ISSN 2229-5518
ARR3-201 transcript of ARR3 gene found in the region be- tween the two different microsatellite markers provided has been analyzed and primers have been designed to amplify its exons. The ARR3 has been selected as a top candidate gene after an intense investigation using different genetic browsers and published articles. Evidence has been provided in the lit- erature that this gene is localized in cone photoreceptors and its associated with different functions in the cones photorecep- tors furthermore the expression rate was significantly high in the retina which provides more evidence that ARR3 gene is possibly associated with cone dystrophy identified within the family.
[1] http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=9606&CHR= X&BEG=67074048&END=70613291&build=37
[2] http://www.ensembl.org/Homo_sapiens/Location/Overview?r=X:672
51011-70436328
[3] http://eyebrowse.cit.nih.gov/cgi- bin/hgTracks?clade=vertebrate&org=Human&db=hg19&position=chr X%3A67251011-
70436328&pix=620&hgsid=183&hgt.dummyEnterButton.x=0&hgt.du mmyEnterButton.y=0
[4] http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=retrieve&d opt=full_report&list_uids=407
[5] http://www.ensembl.org/Homo_sapiens/Transcript/Summary?db=co re;g=ENSG00000120500;r=X:67251011-70436328;t=ENST00000307959
[6] http://www.uniprot.org/uniprot/P36575
[7] http://ihg2.helmholtz-muenchen.de/ihg/ExonPrimer.html
[8] http://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist= Hs.308
IJSER © 2015 http://www.ijser.org