algorithm with time complexity O (
The research paper published by IJSER journal is about An Optimal iterative Minimal Spanning tree Clustering Algorithm for images 1
ISSN 2229-5518
An Optimal iterative Minimal Spanning tree
Clustering Algorithm for images
S. Senthil, A. Sathya, Dr.R.David Chandrakumar
—————————— ——————————
Cluster analysis is playing an important role in solving many problems in medical field, psychology, biology, sociology, pattern recognition and image processing. Due to the limitations in image equipments in MRI, the image has mainly three considerable difficulties: Noise, partial volume and intensity in-homogeneity .Also the image signals have highly affected by shacking of patient’s body and patient’s motion. So the medical MRI is seriously affected and it has improper information about the anatomic structure. Hence the segmentation of medical images is an important one before it to go for treatment planning for proper diagnosis. Automated segmentation methods based on artificial intelligence A spanning tree is an acyclic sub graph of a graph G,
which contains all vertices from G. The minimum spanning tree (MST) of a weighted graph is minimum weight spanning tree of that graph with the classical MST algorithms [2, 3, 4] the cost of constructing a minimum spanning tree is O (mlogn), where m is the number of edges in the graph and n is the number of vertices. More efficient algorithm for constructing MSTs have also been extensively researched [5,
————————————————
1 Department of Mathematics, Vickram College of Engineering, Enathi, Sivagangai, Tamil nadu, India.
2 Department of Mathematics, National Institute of Technology
Goa, Goa Engineering college campus, India.
3 Department of Mathematics, Vickram College of Engineering, Enathi, Sivagangai, Tamil nadu, India.
techniques were proposed in (Clark etal.[1]).The image segmentation viewed as partition a given image into regions(or) segments such that pixels belonging to a region are more similar to each other than pixel belonging to different regions. We also require that these regions be connected so regions consist of neighboring pixels. Image segmentation used to partition a given image into a number of regions. So that each region corresponds to an object(intensity, color, texture…).Here we address the problem of segmenting a digital image into a set of disjoint regions such that each region is composed of nearby pixels with similar colors (or) intensities (or) spatial location.
6, 7].These algorithms promise close to linear time complexity under different assumptions. A Euclidean minimum spanning tree (EMST) is a spanning tree of a set of n points in a metric space ( E n ).where the length of an edge is the Euclidean distance between a pair of points in the point set. MSTs have been used for data Classification in the field of pattern recognition (8) and image processing (9, 10, 11).we have also seen some limited applications in biological data analysis (12).One popular form these MST applications is called the single-linkage cluster analysis (13, 14, 15, 16).
Our study on these methods has led us to believe that
all these applications have used the MSTs in some heuristic ways;eg.cutting long edges to separate clusters without fully exploring their power and understanding their rich properties related to clustering. Geometric notion of centrality are closely linked to facility location problem. The distance matrix D
IJSER © 2012
The research paper published by IJSER journal is about An Optimal iterative Minimal Spanning tree Clustering Algorithm for images 2
ISSN 2229-5518
(data set) can Computed rather efficiently using Dijkstra’s
![]()
algorithm with time complexity O (![]()
![]()
V 2 ln
![]()
V ) (17).
![]()
![]()
![]()
The eccentricity of a vertex in G and radius respectively are defined as![]()
(G) ,
We will use a MST to represent a set of expression data and their significant inter-data relationships to facilitate fast
e (x)
max d(x, y)
y V
and
(G)
min e (x)
x V
rigorous clustering algorithm. Given a point set D in E n
,the
The center of G is the set
hierarchical methods stats by constructing a minimal spanning![]()
C(G) {x![]()
V / e (x)![]()
(G)}
tree(MST) from the points in D.Each edge weight represents
The length of the longest path in the graph is called diameter![]()
the distance ((or)dissimilarity) , u![]()
![]()
v between u and![]()
of the graph G, D(G)
The diameter set of G is
max e(x) .
x V
v,which could be defined as the Euclidean distance ,so we
_
named this MST as EMST1.Next the average weight w of the![]()
Dia(G) {x![]()
V / e(x)![]()
D(G)} .![]()
edges in the entire EMST1 and its standard deviation are
An image pixels represents a mode on vertices and an edge reflects pair wise similarities between the pixels. we take a![]()
_
computed; any edge with w e w![]()
(or)longest edge is
graph-based approach to segmentation. Let G![]()
(V, E)
be an
removed from the tree. This leads to a set of disjoint subtrees
undirected graph with vertices vi![]()
V , the set of elements to![]()
ST {T1 , T2 , T3 ........} .Each of these subtrees
Ti is treated
be segmented and
(vi , v j )
![]()
E corresponding to pairs of
as cluster.
neighboring vertices, each edge![]()
(vi , v j ) E
has a
corresponding weight w(vi , v j ) , which is a non-negative measure of the dissimilarity between neighboring elements v i and v j weights of the edges are computed by a similarity function location, brightness and color. With this
representation, the segmentation task can be solved by
![]()
The MST T into k subtrees {T }k to optimize a more general objective function is given by![]()
k
minimum spanning tree clustering methods.
J (U)
i 1 v Ti
![]()
![]()
v ci ,
In this paper, we will provide in-depth studies for
where c i is the center of
Ti , i![]()
1, 2........k .
MST based clustering. our major contributions include a rigorous formulation for Optimal iterative minimal spanning tree clustering algorithm(OPIMSTCA).we believe it is a good idea to allow users to define their desired similarity within a cluster and allow them to have some flexibility to adjust the
that is to optimize the k-clustering so that the total distance between the ―center‖ of each cluster and its data points is minimized-objective function for data clustering. The centers of clusters are identified using eccentricity of points. These points are a representative point for the each cluster (or)
similarity if the adjustment is needed. In this paper we propose
subtrees. A point
ci is assigned to a cluster i if
optimal iterative minimal spanning tree clustering algorithm![]()
ci Ti , i![]()
1,2....k .The group of center points
c1 , c 2 ......ck
for image segmentation algorithm to address the issues of
undesired clustering structure and unnecessary large number of clusters. Our algorithm works in two phases. The first phase construct the Euclidean distance based MST from the pixels of input image data, then creates subtree (cluster/regions) from minimum spanning tree (MST) by removes the inconsistent edges that satisfy the predefined inconsistence measure. The second phase optimal iterative minimal spanning tree algorithm, which produces optimal (or) best number of clusters with segmentation. The performance of proposed method has been shown with random data for images. Finally experimental results and conclusion we summarize the
strength of our methods and possible improvements.
are connected and again minimum spanning tree EMST2 is constructed. To each Ti calculate standard deviation, distance between the point of Ti and clusters center ci .Thus the problem of finding the optimal number of clusters of a data set can be transformed into problem of finding the proper region (clusters) of the data set. Here we use the MST as a criterion to test the inter –cluster property based on this observation, we use a cluster validation criterion, called cluster separation (CS) in OPIMST clustering algorithm.
Cluster separation :(CS) is defined as the ratio between
minimum and maximum standard deviation of clusters
(subtrees),
IJSER © 2012
The research paper published by IJSER journal is about An Optimal iterative Minimal Spanning tree Clustering Algorithm for images 3
ISSN 2229-5518
![]()
CS i , i
i
![]()
1,2........k ,![]()
_
9. If (w e w![]()
) (or) current longest edge e remove e1![]()
where i is the maximum value of standard deviation of
from EMST1.![]()
10. ST![]()
ST {T ' }// T ' is new disjoint
clusters and i is the minimum value of standard deviation
subtrees (regions).
clusters.
Then the CS represents the relative separation of centroids.The
11. n c![]()
![]()
n c 1 .
value of CS ranges from 0 to 1.A low value of CS means that the two centroids are too close to each other and the
12. Compute the center
points.
ci of
Ti using eccentricity of
corresponding partition is not valid. A high CS value means
13. C![]()
Ti
![]()
ST {c i } .
the partitions of the data is even and valid. In practice, we
predefine a threshold to test the CS.If the CS is greater than the threshold; the partition of the data set is valid. Then again
14. Construct an EMST2 T from C.
15. Compute the standard deviation![]()
( Ti ) .
partitions the data set by creating subtree (cluster/region).This process continuous until the CS is smaller than the threshold
The CS criterion finds the proper binary relationship among
16.
17.![]()
( Ti ) =get-min standard deviation.![]()
![]()
( Ti ) =get-max standard deviation.![]()
( Ti )
clusters in the data space. The value setting of the threshold for the CS will be practical and is dependent on the dataset.
The high the value of the threshold the smaller the number of
18. CS=
19. Until CS
, i
(Ti )![]()
0.8 .
1,2......k
clusters would be Generally, the value of the threshold will be
20. Merge the closest neighbor from EMST2.![]()
0.8 [18].The given clusters the CS value
![]()
0.8
and our
21. Update the clusters points, repeat step 12 to step 20.
22. The following optimize the k-clustering objective
OPIMST algorithm processing the results, the proper number
of clusters/regions for the data set (pixel) is 2.Further more,
function minimized, termination criterion is satisfied![]()
the computational cost of CS is much lighter because the
J t (U)![]()
![]()
J t 1 (U)![]()
, where t is the iteration count
number of sub clusters is small. The created clusters/regions are well separated.
Input : Image data (pixel value) Output : optimal number of clusters
Let e1 be an edge in the EMST1 constructed from image data.
Let e2 be an edge in the EMST2 constructed form C.![]()
and is a thresholding value lies between 0 and 1.
This section describes some experimental results on random data to the segmentation Performance of proposed method,
Let w e
be the weight of e1.
Let ![]() be the standard deviation of the edge weights in
be the standard deviation of the edge weights in
EMST1.
Let ST be the set of disjoint subtrees of EMST1.
1. Create a node v, for each pixel of an image I.
2. Compute the edge weight using Euclidean distance from image data.
3. Construct an EMST1 from 2
4. Compute the average weight of from EMST1.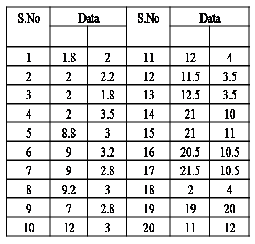
_
w of all the edges
5. Compute the standard deviation ![]() of the edges from
of the edges from
EMST1.
6. ST ![]() , n c
, n c
![]()
![]()
1 , C .![]()
7. Repeat.
8. For each e1
EMST1 .
IJSER © 2012
The research paper published by IJSER journal is about An Optimal iterative Minimal Spanning tree Clustering Algorithm for images 4
ISSN 2229-5518
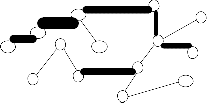
from the EMST1,vertices(data points) in the EMST1 partitioned into four sets (four subtrees (or) clusters)
T1 , T2 , T3 and T4 namely![]()

T1 {1,2,3,4,5,6,7,8,9,10,11,12,13,18,20} , T2![]()
{14,15,16 ,17} ,![]()
T3 {19}
and T4![]()
{20} is show in the figure 2.center point
(vertex) for each of the each subtree is find using eccentricity of points (vertices).These center point (or) vertex is connected and again another minimum spanning tree EMST2(figure3) is constructed. To calculate the standard deviation each subtree using center. The maximum value of standard deviation of clusters ![]() i and the minimum value of standard deviation of clusters
i and the minimum value of standard deviation of clusters ![]() i , is find to compute cluster separation value. If the CS is greater than 0.8, then to remove the minimum edge weight of EMST2 .To update the clusters(subtree) vertices then to compute the center using eccentricity points(vertices).Finally the optimal iterative minimal spanning tree clustering algorithm produce, the optimal number of clusters 2.
i , is find to compute cluster separation value. If the CS is greater than 0.8, then to remove the minimum edge weight of EMST2 .To update the clusters(subtree) vertices then to compute the center using eccentricity points(vertices).Finally the optimal iterative minimal spanning tree clustering algorithm produce, the optimal number of clusters 2.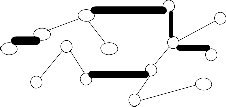
T4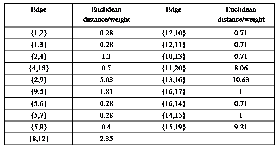
8
12 7
8
12 7
10
5
13 1 11
6
10
5
13 1 11
6
9
3 2 18
4
9
3 2 18 T

4
17
16
14
Our OPIMSTCA constructs EMST1 from the dissimilarity 15
_
matrix is shown in the figure1.The mean
w and standard
deviation ![]() of the edges from the EMST1 are computed
of the edges from the EMST1 are computed
_
respectively as 2.38 and 4.66.The sum of the mean
w and T3 T4
standard deviation ![]() is computed as 7.04.This value is used to identify the inconsistence edges in the EMST1 to generate subtrees(clusters).based on the above value the edge having
is computed as 7.04.This value is used to identify the inconsistence edges in the EMST1 to generate subtrees(clusters).based on the above value the edge having
weight 8.06,9.21 and 10.63.By removing inconsistent edges![]()
![]()
19 20
IJSER © 2012
The research paper published by IJSER journal is about An Optimal iterative Minimal Spanning tree Clustering Algorithm for images 5
ISSN 2229-5518

5
20
19
14
Our algorithm OPIMST clustering algorithm, processing the optimal number of clusters Two.
generalization‖, Bell systems technical journal
36:1389- 1401(1957).
[3].Kruskal.J. ―On the Shortest spanning subtree and the travelling salesman problem ―In proceedings of the American Mathematical Society, Pages
48-50(1956).
[4].Nesetril.J, Milkova.E and Nesetrilova.H.otakar boruvka―On minimal Spanning tree problem:‖Translation of both the 1926 papers, comments, history.DMATH‖.Discrete Mathematics, 233(2001).
[5].Karger.D, Klein.P and Tarjan.R ―A randomized linear-time algorithm to find minimum spanning tree‖, Journal of the ACM, 42(2):321-
328(1995).
[6].Fredman.M and Willard.D ―Trans-dichotomous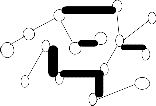
17
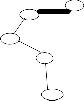
16 12
10
14
13 1 11
15
3 2
19
8
7
20 5
6
9
18
4
algorithms for minimum spanning trees and shortest paths‖ ,In proceedings of the 31st annual IEEE symposium on Foundations of computer science,pages 719-725(1990).
[7].Gabow.h,Spencer.T and Rarjan.R ,‖Efficient
algorithms for finding minimal spanning trees in undirected and directed graphs‖, Combinatorica
6(2):109-122(1986).![]()
i 2.94 and
![]()
i 3.60 CS
![]()
0.82![]()
0.8
[8].Duda.R.O and hart .P.E. ―pattern classification
and scene analysis‖ wiley-inter Sceince, New
In this paper proposed an optimal iterative MST clustering algorithm and applied to random data segmentation. The algorithm was formulated by introducing Euclidean distance function, eccentricity, and standard deviation of each cluster, with basic Objective function of the optimal iterative minimal spanning tree clustering algorithm to have proper effective segmentation in image. Our algorithm uses new cluster validation criterion based on the geometric property of partitioned clusters to produce optimal number of true clusters. The test result shows that the proposed method outperformed the base line methods. In future we will explore and test our proposed clustering algorithm in various domains and this work hopes that the proposed method can also be used to improve the performance of other clustering algorithm based on Euclidean distance functions.
References: [1].Clark,M.C.,Hall,L.O.,Goldgof,D.B.,Velthuizen,R.
Murtagh,F.R.,Silbiger,M.S:- ―Automatic tumor- segmentation using knowledge-based technique‖. IEEE Transactions on Medical Imaging 117,187-
201(1998).
[2].Prim.R. ―Shortest connection networks and some
York (1973).
[9].Gonzalez.R.C and wintz.P ―Digital image processing‖, 2nd edn, Addison-wesley., Reading MA (1987).
[10].Xu, Y.Olman.V and Uberbacher.E. ―A segmentation algorithm for noisy images; design and evaluation‖, patt.recogn.lett19, 1213-1224
C (1998).
[11].Xu.y and Uberbacher.E. ―2D image segmentation using minimum spanning trees‖
,image Vis.comput 15, 47-57(1997).
[12].States.D.JHarris, N.L.and Hunter,
―Computationally efficient cluster representation in molecula Sequence megaclassification‖,
Ismb, 1,387-394(1993).
[13].Gower J.C and Ross .G.J.S ―minimum spanning
trees and single linkage analysis‖, Appl.stat.18,
54-64(1969).
[14].Aho.A.V, Hopcroff.J.E and Ullman.J.D, ―The Design and Analysis of computer algorithms‖, Addison- wesley, Reading MA (1974).
[15].A.k and Dubes .R. ―Algorithms for clustering
Data‖, prentice –hall, New Jersey (1988).
IJSER © 2012
The research paper published by IJSER journal is about An Optimal iterative Minimal Spanning tree Clustering Algorithm for images 6
ISSN 2229-5518
[16].Mirkin.B, ―Mathematical classification and clustering –DIMACS‖, Rutgers University, Piscataway.Nj (1996).
[7].Stefan wuchty and peter .F.Stadler, ―Centers of
complex networks‖(2006).
[18].FengLuo,Latifur kahn,Farokh Bastani,T-ling yen and Jizhong zhon, ―A dynamically growing self- organizing tree(DGOST)for hierarchical gene expression profile‖,Bio informatics,Vol20,No
16,PP2605-2617,(2004).
Authors:
1. S.Senthil is working as Assistant professor in Mathematics, Vickram College of Engineering, Enathi, Sivagangai.He earned his M.Sc degree from Saraswathi Narayanan College, Madurai Kamaraj University, Madurai, He also earned his M.Phil from Saraswathi Narayanan College, Madurai Kamaraj University, Madurai.Now he is doing Ph.D in Mathematics at Anna University of Technology Madurai, Madurai.Email:senthil.lmec@gmail.com.
2. A.sathya is working as Assistant professor in Mathematics, National Institute of Technology Goa; Goa.she earned her M.Sc degree from Saraswathi Narayanan College, Madurai Kamaraj University, and Madurai. She also earned her M.Phil from Vinayaka missions University Selam. She was published research papers on clustering algorithm in various international journals. She was submitted Ph.D thesis on ―Fuzzy Clustering Analysis in Medical images‖ at Gandhigram University, Dindigul. Email:sathyaarumugam.gru@gmail.com
3. R.David Chandrakumar received his graduate degree in Mathematics from M.D.T. Hindu College, Tirunelveli in 1969.Post graduate degree in Mathematics from St.Xavier's College, Tirunelveli in 1972.He received M.Phil in Mathematics from University of Jammu in 1980.He also Received Ph.D in Mathematics from University of Jammu in 1986.Presently he is working as a Professor in Mathematics department of Vickram College of Engineering, Enathi, Sivagangai Email:mathsvce@gmail.com.
IJSER © 2012