International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 1
ISSN 2229-5518
Algorithm of Insulin Human P01308 – Discrete Code 2029
Lutvo Kurić
Abstract - The modern science mainly treats the biochemical basis of sequencing in bio-macromolecules and processes in medicine and biochemistry. One can ask weather the language of biochemistry is the adequate scientific language to explain the phenomenon in that science. Is there maybe some other language, out of biochemistry, that determines how the biochemical processes will function and what the structure and organization of life systems will be? The research results provide some answers to these questions. They reveal to us that the process of sequencing in bio-macromolecules is conditioned and determined not only through biochemical, but also through cybernetic and information principles. Many studies have indicated that analysis of protein sequence codes and various sequence-based prediction approaches, such as predicting drug-target interaction networks (He et al., 2010), predicting functions of proteins (Hu et al., 2011; Kannan et al., 2008), analysis and prediction of the metabolic stability of proteins (Huang et al., 2010), predicting the network of substrate-enzyme-product triads (Chen et al., 2010), membrane protein type prediction (Cai and Chou, 2006; Cai et al., 2003; Cai et al., 2004), protein structural class prediction (Cai et al., 2006; Ding et al., 2007), protein secondary structure prediction (Chen et al., 2009; Ding et al., 2009b), enzyme family class prediction (Cai et al., 2005; Ding et al., 2009a; Wang et al., 2010), identifying cyclin proteins (Mohabatkar, 2010), protein subcellular location prediction (Chou and Shen, 2010a; Chou and Shen, 2010b; Kandaswamy et al., 2010; Liu et al., 2010), among many others as summarized in a recent review (Chou, 2011), can timely provide very useful information and insights for both basic research and drug design and hence are widely welcome by science community. The present study is attempted to develop a novel sequence-based method for studying insulin in hopes that it may become a useful tool in the relevant areas.
Index Terms-Discrete Code, Human Insulin, Insulin Model, Insulin Code.
—————————— ——————————
HE biologic role of any given protein in essential life processes, eg, insulin, depends on the positioning of its component amino acids, and is understood by the „positioning of letters forming words“. Each of these words has its biochemical base. If this base is expressed by corresponding discrete numbers, it can be seen that any given base has its own program, along with
its own unique cybernetics and information characteristics.
Indeed, the sequencing of the molecule is determined not only by distin biochemical features, but also by cybernetic and information principles. For this reason, research in this field deals more with the quantitative rather than qualitative characteristcs of genetic information and its biochemical basis. For the purposes of this paper, specific physical and chemical factors have been selected in order to express the genetic information for insulin. Numerical values are them assigned to these factors, enabling them to be measured. In this way it is possible to determine oif a connection really exists between the
quantitative ratios in the process of transfer of genetic information and the qualitative appearance of the insulin molecule. To select these factors, preference is given to classical physical and chemical parameters, including the number of atoms in the relevant amino acids, their analog values, the position in these amino acids in the peptide chain, and their frenquencies. There is a arge numbers of these parameters, and each of their gives important genetic information. Going through this process, it becomes clear that there is a mathematical relationship between quantitative ratios and the qualitative appearance of the biochemical „genetic processes“ and that there is a measurement method that can be used to describe the biochemistry of insulin.
The biologic role of any given protein in essential life processes, eg, insulin, depends on the positioning of its component amino acids, and is understood
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 2
ISSN 2229-5518
by the „positioning of letters forming words“. Each of these words has its biochemical base. If this base is expressed by corresponding discrete numbers, it can be seen that any given base has its own program, along with its own unique cybernetics and information characteristics. Indeed, the sequencing of the molecule is determined not only by distin biochemical features, but also by cybernetic and information principles. For this reason, research in this field deals more with the quantitative rather than qualitative characteristcs of genetic information and its biochemical basis. For the purposes of this paper, specific physical and chemical factors have been selected in order to express the genetic information for insulin. Numerical values are them assigned to these factors, enabling them to be measured. In this way it is possible to determine oif a connection really exists between the quantitative ratios in the process of transfer of genetic information and the qualitative appearance of the insulin molecule. To select these factors, preference is given to classical physical and chemical parameters, including the number of atoms in the relevant amino acids, their analog values, the position in these amino acids in the peptide chain, and their frenquencies. There is a arge numbers of these parameters, and each of their gives important genetic information. Going through this process, it becomes clear that there is a mathematical relationship
between quantitative ratios and the qualitative appearance of the biochemical
„genetic processes“ and that there is a measurement method that can be used
to describe the biochemistry of insulin.
Insulin can be represented by two different forms, ie, a discrete form and a sequential form. In the discrete form, a molecule of insulin is represented by a set of discrete codes or a multiple dimension vector. In the sequential form, an insulin molecule is represent by a series of amino acids according to the order of their position in the sequence lenght 110 AA.
Therefore, the sequential form can naturally reflect all the information about the sequence order and lenght of an insulin molecule. The key issue is whether we can develop a different discrete method of representing an insulin molecule that will allow accomodation of partial, if not all sequence order information? Because a protein sequence is usually represented by a series of amino acids should be assigned to these codes in order to optimally convert the sequence order information into a series of numbers for the discrete form representation?
The matrix mechanism of Insulin, the evolution of biomacromolecules and, especially, the biochemical evolution of Insulin language, have been analyzed by the application of cybernetic methods, information theory and system theory, respectively. The primary structure of a molecule of Insulin is the exact specification of its atomic composition and the chemical bonds connecting those atoms.
M | A | L | W | M | R | L | L | P | L | L | A | L | L | A | L | W | G | P | D | P | A |
A | A | F | V | N | Q | H | L | C | G | S | H | L | V | E | A | L | Y | L | V | C | G |
E | R | G | F | F | Y | T | P | K | T | R | R | E | A | E | D | L | Q | V | G | Q | V |
E | L | G | G | G | P | G | A | G | S | L | Q | P | L | A | L | E | G | S | L | Q | K |
R | G | I | V | E | Q | C | C | T | S | I | C | S | L | Y | Q | L | E | N | Y | C | N |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 3
ISSN 2229-5518
We shall now give some mathematical evidences that will prove that in the biochemistry of insulin in there really is programmatic and cybernetic algorithm in which it is „recorded“, in the language of mathematics, how the molecule will be built and what will be the quantitative characteristics of the given genetic information.
[AC1 + (AC1+ AC2) + (AC1+ AC2+ AC3)..., + (AC1+ AC2+ AC3..., + ACR)] = S1; AC1 = APa1;
(AC1+ AC2) = APa2 ;
(AC1+ AC2+ AC3) = APa3; (AC1+ AC2+ AC3..., + AC306) = APaR;
APa1,2,3,n = Atomic progression of amino acids 1,2,3,n
[APa1+APa2+APa3)..., + APaR)] = S1;
[ACR + (ACR+ AC(R-1)) + (ACR+ AC(R-1)+ Ac(R-2))..., + (ACR+AC(R-1)+AC(R-2)..., +AC1)] = S2; ACR = APbR;
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 4
ISSN 2229-5518
(ACR+ AC(R-1)) = APbR; (ACR+ AC(R-1)+ AC(R-2)) = APb(R-2);
(ACR+ AC(R-1)+ AC(R-2)..., + AC1) = APb1;
APbR,(R-1),(R-2), …,n = Atomic progression of amino acids R,(R-1),(R-2),…n;
[APbR+APb(R-1)+APb1(R-2))..., + APb1)] = S2;
Progressions can be: macro and micro, even and odd, primary and secondary, analogue, negative, positive, etc.
Cybernetic, information and system characteristics of biochemistry of insulin can be researched also using frequencies (macro and micro), primary and secondary values, standard deviations, analogue values, even and odd values, determinants, bio codes, etc.
Within the digital pictures in biochemistry, the physical and chemical parameters are in a strict compliance with programmatic, cybernetic and information principles. Each bar in the protein chain attracts only the corresponding aminoacid, and only the relevant aminoacid can be positioned at certain place in the chain. Each peptide chain can have the exact number of aminoacids necessary to meet the strictly determined mathematical conditioning. It can have as many atoms as necessary to meet the mathematical balance of the biochemical phenomenon at certain mathematical level, etc. The digital language of biochemistry has a countless number of codes and analogue codes, as well as other information content. These pictures enable us to realize the very essence of functioning of biochemical processes. There are some examples:
Number of atoms Position
L L L L L L L L L L L
AA 3 7 8 10 11 13 14 16 30 35 39 41 61 68 77 80 82 86 102 105
APa 55 150 172 211 233 268 290 325 572 652 725 771 1155 1284 1400 1459 1494 1559 1850 1916
![]()
APb 1974 1879 1857 1818 1796 1761 1739 1704 1457 1377 1304 1258 874 745 629 570 535 470 179 113
AP(a,b) 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029 2029
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 5
ISSN 2229-5518
Notes: Namely, having mathematically analyzed the atomic progression model of Insulin (Table 1) we have found out that the protein code is based on a periodic law. This being the only to „read“ the picture, the solution of the main problem (concering an arrangement where each amino acid takes only one, precisely determined position in the code), is quite manifest.
When evaluating progressions, one has to take into account the fact that there are macro and micro progressions, odd and even, primary and secondary, analogue, etc. Progressions have their category (odd and even, primary and secondary, analogue, etc.) All these progressions are in correlation with each other.
Establishing of numeric values of amino acids needs to be done through use of strictly determined criterion from the theory of systems and also from cybernetics whish, in this example, is the number of atoms in amino acids. That is only one dimension of the digital image of insulin. There are many other dimensions as well as digital images. Each of these dimensions and images has its corresponding progression. With some dimensions, one has to use some other parameters from the theory of systems and cybernetics (frequency, standard deviation, various codes and analogue codes, analogue values, primary and secondary values, odd-even relation, and many others), and not progression.
Regardless of the fact whether there is a typical correlation between parameters or not, their effect in the process of evolution can be followed through use of adequate methodology.
In digital pictures of biochemistry, physical and chemical parameters are in a strict submission to programmed, cyber and information rules. In some examples, chemical elements are connected through the discrete codes 19 and 7, which is transformed into 2029 code. That code we can find using the following algorithm:
{(SB(X1,2,3,n) x A ) - (SA(X1,2,3,n) x B ) + (AB)} = ABA;
{(S7(X1,2,3,n) x A ) - (S19(X1,2,3,n) x B ) + (AB)} = (19x7x19);
S = Groups of amino acids 1,2,3,n
X1,2,3,n = Number of atoms
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 6
ISSN 2229-5518
Connection is one of numerical expressions that connects various corresponding features in biochemistry. It has a very prominent place in the mathematical picture of all processes in biochemistry. Here are some examples:
Codes 19 and 7 197; Codes 7 and 19 719;
Discrete codes 19 and 7, is transformed into 2029 code.
We shall now give some mathematical evidences that will prove that in the biochemistry there really is programmatic and cybernetic algorithm in which it is „recorded“, in the language of mathematics, how the molecule will be built and what will be the quantitative characteristics of the given biochemical information. Dicsret codes 19 and 7 is an area of bio-macromolecules and processes in biochemistry (chemical engineering, bioprocess engineering, information technology, biorobotics) that treats signals as stochastic processes, dealing with their biosignal properties (e.g., frequences, mean, covariance, etc.). In this context those codes are modeled as functions consisting of both deterministic and stochastic components. A simple example and also a common model of many bio systems is a code 2029 that consists of a deterministic part x(t) as white this code.![]()
The mathematical balance in groups of chemical elements from X to Y on exsist with help codes 19 i 7. Output those codes is code 2029.
AA1 AA2 AA3 . . An
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 7
ISSN 2229-5518
Na Na Na . . Na
![]()
![]()
AP1 AP2 AP3 . . Apn
. .
DP1 DP2 DP3 . . DPn > Discret code | 2029 | |||||||||
| | | . | . | | | | | | |
APa,b | AP1 AP2 AP3 . . Apn |
P1 | P2 | P3 | . | . | Pn |
AA1,2,3,n = Amino acids 1,2,3,n
Na = Number of atoms P1,2,3,n = Position of amino acids APa,b = Atomic progression a,b, DP1,2,3,n = Progression differences
The subject of the research we are discussing in this text is the cyber-information access to the research of the amino acidic constitution of insulin. Strictly speaking, the subject of this research is finding of an adequate scientific language that could describe this phenomenon, study of the genetic information, as well as relationship between the genetic language of the protein and the theory of the system and cybernetics.
We shall also seek the answers for the following questions: Does the matrix mechanism of biosynthesis of this protein function within the law of the general theory of information and theory of system, and what is the significance of it for understanding of the genetic language of insulin? What is the essence of existence and functioning of this language? Is the genetic information characterized only by biochemical, or also by cyber- information principles? Etc...
We shall also analyze potential effects of physical and chemical and cybernetic and information principles in the biochemical base of insulin. Biological specificum of protein, its place and role in life processes depends on the positioning of amino acids in its molecules.
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 8
ISSN 2229-5518
The molecule of insulin we can understand as words built from letters, i.e. aminoacids. The meaning of words is determined by positioning of letters. Each of these words has its biochemical base. If this base is expressed by corresponding discrete numbers, we find out that the base has its own program, cybernetic and information characteristics. In fact, we will find out that the sequencing of the molecule is conditioned and determined not only by biochemical, but also by cybernetic and information principles.
For this reason, in this research we will deal more with quantitative, and less with qualitative characteristics of the genetic information and its biochemical foundation.
Progression APa
105 105 105 105 105 105 105 105 105 105
![]()
1916 1916 1916 1916 1916 1916 1916 1916 1916 1916
1861 1766 1744 1705 1683 1648 1626 1591 1344 1264 > (2029 x Y)
![]()
55 150 172 211 233 268 290 325 572 652
L L L L L L L L L L
22 22 22 22 22 22 22 22 22 22
3 7 8 10 11 13 14 16 30 35
(1916 – 55) = 1861; (1916-150) = 1766; (1916-172) = 1744; etc.
(1861+1766+1744…+ 1264) = (2029 + 2029 + 2029…, + 2029):
Y = 8;
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 9
ISSN 2229-5518
Progression APb
105 105 105 105 105 105 105 105 105 105
![]()
113 113 113 113 113 113 113 113 113 113
| | | | | | | | | | ||
-1861 | -1766 | -1744 | -1705 | -1683 | -1648 | -1626 | -1591 | -1344 | -1264 | > | (-)2029 x Y) |
![]()
1974 1879 1857 1818 1796 1761 1739 1704 1457 1377
L L L L L L L L L L
22 22 22 22 22 22 22 22 22 22
3 7 8 10 11 13 14 16 30 35
Table 2. Schematic representation of the atomic progression APa APb (Amino acid Leu – position from discret code 2029).
APb | APa | APb | APa | APb | APa | |||||||
L | L | L | L | L | L |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 10
ISSN 2229-5518
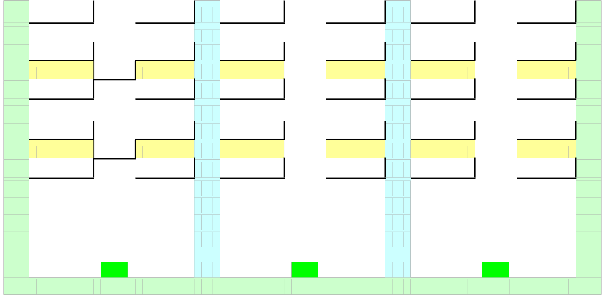
Determinants 2x2
1916 | 55 | > | (1861 x 2029) |
113 | 1974 |
Determinants 2x2
1916 | 150 | > | (1766 x 2029) |
113 | 1879 |
Determinants 2x2
1916 | 172 | > | (1744 x 2029) |
113 | 1857 |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 11
ISSN 2229-5518
AP-b AP-a AP-b AP-a AP-b AP-a
L | L | L | L | L | L | ||||||||||
22 | 22 | 22 | 22 | 22 | 22 | ||||||||||
105 | 105 | 105 | 105 | 105 | 105 | ||||||||||
| | | | | | ||||||||||
113 | 1916 | 113 | 1916 | 113 | 1916 | ||||||||||
| | | | | | ||||||||||
-1705 | 1705 | -1683 | 1683 | -1648 | 1648 | ||||||||||
| | | | | | ||||||||||
1818 | 211 | 1796 | 233 | 1761 | 268 | ||||||||||
| | | | | | ||||||||||
L | L | L | L | L | L | ||||||||||
22 | 22 | 22 | 22 | 22 | 22 | ||||||||||
10 | 10 | 11 | 11 | 13 | 13 | ||||||||||
DET | DET | DET | |||||||||||||
2029 | 2029 | 2029 |
Determinants 2x2 | |||
1916 | 211 | > | (1705 x 2029) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 12
ISSN 2229-5518
113 1818
Determinants 2x2
Determinants 2x2 | |||
1916 | 290 | > | (1626 x 2029) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 13
ISSN 2229-5518
113 1739
Determinants 2x2
1916 | 325 | > | (1591 x 2029) |
113 | 1704 |
Determinants 2x2
1916 | 572 | > | (1344 x 2029) |
113 | 1457 |
Determinants 2x2
1916 | 652 | > | (1264 x 2029) |
113 | 1377 |
Table 3. Schematic representation of the atomic progression APa APb (Amino acid Leu – position from discret code 2029).
Atomic progressions in correlation with each other result in the progression difference, and the progression difference result in the discrete code
2029. We could say that the 2029 code connects all the progressions into the progression matrix Ap-a and Ap-b. That code connects the progressions with the number of atoms in insulin.
14 | 16 | 30 | 35 | 39 | 82 | > | 216 | |
AP-a | 290 | 325 | 572 | 652 | 725 | 1494 | > | (2029 x 2) |
14 | 16 | 30 | 35 | 39 | 82 | > | 216 | |
AP-b | 1739 | 1704 | 1457 | 1377 | 1304 | 535 | > | (2029 x 4) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 14
ISSN 2229-5518
10 | 11 | 41 | 68 | 86 | > | 216 | |
AP-a | 211 | 233 | 771 | 1284 | 1559 | > | (2029 x 2) |
10 | 11 | 41 | 68 | 86 | > | 216 | |
AP-b | 1818 | 1796 | 1258 | 745 | 470 | > | (2029 x 3) |
10 | 11 | 13 | 39 | 41 | 102 | > | 216 | |
AP-a | 211 | 233 | 268 | 725 | 771 | 1850 | > | (2029 x 2) |
10 | 11 | 13 | 39 | 41 | 102 | > | 216 | |
AP-b | 1818 | 1796 | 1761 | 1304 | 1258 | 179 | > | (2029 x 4) |
8 | 13 | 16 | 35 | 39 | 105 | > | 216 | |
AP-a | 172 | 268 | 325 | 652 | 725 | 1916 | > | (2029 x 2) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 15
ISSN 2229-5518
8 | 13 | 16 | 35 | 39 | 105 | > | 216 | |
AP-b | 1857 | 1761 | 1704 | 1377 | 1304 | 113 | > | (2029 x 4) |
7 | 14 | 16 | 35 | 39 | 105 | > | 216 | |
AP-a | 150 | 290 | 325 | 652 | 725 | 1916 | > | (2029 x 2) |
7 | 14 | 16 | 35 | 39 | 105 | > | 216 | |
AP-b | 1879 | 1739 | 1704 | 1377 | 1304 | 113 | > | (2029 x 4) |
3 | 8 | 13 | 14 | 35 | 41 | 102 | > | 216 | |
AP-a | 55 | 172 | 268 | 290 | 652 | 771 | 1850 | > | (2029 x 2) |
3 | 8 | 13 | 14 | 35 | 41 | 102 | > | 216 | |
AP-b | 1974 | 1857 | 1761 | 1739 | 1377 | 1258 | 179 | > | (2029 x 5) |
![]()
L L L L L L L Sum
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volum | e 2, Issue 8, August-2011 | 16 |
ISSN 2229-5518 | ||
22 22 22 22 | 22 22 22 | |
3 8 11 16 35 41 102 | > 216 | |
AP-a | 55 172 233 325 652 771 1850 | > (2029 x 2) |
![]()
3 | 8 | 11 | 16 | 35 | 41 | 102 | > | 216 | |
AP-b | 1974 | 1857 | 1796 | 1704 | 1377 | 1258 | 179 | > | (2029 x 5) |
8 | 11 | 13 | 41 | 61 | 80 | > | 214 | |
AP-a | 172 | 233 | 268 | 771 | 1155 | 1459 | > | (2029 x 2) |
8 | 11 | 13 | 41 | 61 | 80 | > | 214 | |
AP-b | 1857 | 1796 | 1761 | 1258 | 874 | 570 | > | (2029 x 4) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 17
ISSN 2229-5518
AP-a | 172 | 211 | 325 | 572 | 1284 L | 1494 L | > | (2029 x 2) Sum |
8 | 10 | 16 | 30 | 68 | 82 | > | 214 | |
AP-b | 1857 | 1818 | 1704 | 1457 | 745 | 535 | > | (2029 x 4) |
L | Sum | |||||||
8 | 10 | 14 | 41 | 61 | 80 | > | 214 | |
AP-a | 172 | 211 | 290 | 771 | 1155 | 1459 | > | (2029 x 2) |
L | Sum | |||||||
8 | 10 | 14 | 41 | 61 | 80 | > | 214 | |
AP-b | 1857 | 1818 | 1739 | 1258 | 874 | 570 | > | (2029 x 4) |
L | Sum | |||||||
7 | 13 | 16 | 30 | 68 | 80 | > | 214 | |
AP-a | 150 | 268 | 325 | 572 | 1284 | 1459 | > | (2029 x 2) |
L | Sum | |||||||
7 | 13 | 16 | 30 | 68 | 80 | > | 214 | |
AP-b | 1879 | 1761 | 1704 | 1457 | 745 | 570 | > | (2029 x 4) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 18
ISSN 2229-5518
7 | 13 | 14 | 30 | L 68 | L 82 | > | Sum 214 | |
AP-a | 150 | 268 | 290 | 572 | 1284 | 1494 | > | (2029 x 2) |
L | Sum | |||||||
7 | 13 | 14 | 30 | 68 | 82 | > | 214 | |
AP-b | 1879 | 1761 | 1739 | 1457 | 745 | 535 | > | (2029 x 4) |
L | Sum | |||||||
7 | 11 | 16 | 30 | 68 | 82 | > | 214 | |
AP-a | 150 | 233 | 325 | 572 | 1284 | 1494 | > | (2029 x 2) |
L | Sum | |||||||
7 | 11 | 16 | 30 | 68 | 82 | > | 214 | |
AP-b | 1879 | 1796 | 1704 | 1457 | 745 | 535 | > | (2029 x 4) |
L | Sum | |||||||
7 | 11 | 14 | 41 | 61 | 80 | > | 214 | |
AP-a | 150 | 233 | 290 | 771 | 1155 | 1459 | > | (2029 x 2) |
L | Sum | |||||||
7 | 11 | 14 | 41 | 61 | 80 | > | 214 | |
AP-b | 1879 | 1796 | 1739 | 1258 | 874 | 570 | > | (2029 x 4) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 19
ISSN 2229-5518
3 | 8 | 11 | 13 | 16 | 61 | L 102 | > | Sum 214 | |
AP-a | 55 | 172 | 233 | 268 | 325 | 1155 | 1850 | > | (2029 x 2) |
Sum | |||||||||
AP-b | 3 | 8 | 11 | 13 | 16 | 61 | 102 | > | 214 |
1974 | 1857 | 1796 | 1761 | 1704 | 874 | 179 | > | (2029 x 5) |
etc.
AP-a 290 325 652 771 1155 1400 1494 > (2029 x 3)
AP-b 1739 1704 1377 1258 874 629 535 > (2029 x 4)
AP-a 233 572 652 725 771 1284 1850 > (2029 x 3)
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 20
ISSN 2229-5518
L | L | |||||
22 | 22 | 22 | 22 | 22 | 22 | 22 |
11 | 30 | 35 | 39 | 41 | 68 | 102 | > | 326 | |
AP-b | 1796 | 1457 | 1377 | 1304 | 1258 | 745 | 179 | > | (2029 x 4) |
L | L | L | L | |||
22 | 22 | 22 | 22 | 22 | 22 | 22 |
(2029 x 3)
(2029 x 4)
3 | 7 | 10 | 16 | 35 | 68 | 82 | 105 | > | 326 | |
AP-a | 55 | 150 | 211 | 325 | 652 | 1284 | 1494 | 1916 | > | (2029 x 3) |
L | L | Sum | ||||||||
3 | 7 | 10 | 16 | 35 | 68 | 82 | 105 | > | 326 | |
AP-b | 1974 | 1879 | 1818 | 1704 | 1377 | 745 | 535 | 113 | > | (2029 x 5) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 21
ISSN 2229-5518
3 | 7 | 10 | 11 | 13 | 41 | 80 | 82 | 86 | 105 | > | 438 | |
AP-a | 55 | 150 | 211 | 233 | 268 | 771 | 1459 | 1494 | 1559 | 1916 | > | (2029 x 4) |
L | L | L | Sum | |||||||||
3 | 7 | 10 | 11 | 13 | 41 | 80 | 82 | 86 | 105 | > | 438 | |
AP-b | 1974 | 1879 | 1818 | 1796 | 1761 | 1258 | 570 | 535 | 470 | 113 | > | (2029 X 6) |
L | L | L | L | Sum | ||||||||
3 | 8 | 14 | 16 | 30 | 35 | 39 | 86 | 102 | 105 | > | 438 | |
AP-a | 55 | 172 | 290 | 325 | 572 | 652 | 725 | 1559 | 1850 | 1916 | > | (2029 x 4) |
L | L | L | L | Sum | ||||||||
3 | 8 | 14 | 16 | 30 | 35 | 39 | 86 | 102 | 105 | > | 438 | |
AP-b | 1974 | 1857 | 1739 | 1704 | 1457 | 1377 | 1304 | 470 | 179 | 113 | > | (2029 X 6) |
L | L | L | Sum | |||||||||
3 | 10 | 11 | 13 | 14 | 39 | 77 | 80 | 86 | 105 | > | 438 | |
AP-a | 55 | 211 | 233 | 268 | 290 | 725 | 1400 | 1459 | 1559 | 1916 | > | (2029 x 4) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 22
ISSN 2229-5518
3 | 10 | 11 | 13 | 14 | 39 | 77 | 80 | 86 | 105 | > | 438 | |
AP-b | 1974 | 1818 | 1796 | 1761 | 1739 | 1304 | 629 | 570 | 470 | 113 | > | (2029 X 6) |
3 | 10 | 13 | 35 | 41 | L 68 | L 77 | L 86 | L 105 | > | Sum 438 | |
AP-a | 55 | 211 | 268 | 652 | 771 | 1284 | 1400 | 1559 | 1916 | > | (2029 x 4) |
L | L | L | L | Sum | |||||||
3 | 10 | 13 | 35 | 41 | 68 | 77 | 86 | 105 | > | 438 | |
AP-b | 1974 | 1818 | 1761 | 1377 | 1258 | 745 | 629 | 470 | 113 | > | (2029 X 5) |
L | L | L | L | Sum | |||||||
3 | 14 | 16 | 35 | 41 | 61 | 80 | 86 | 102 | > | 438 | |
AP-a | 55 | 290 | 325 | 652 | 771 | 1155 | 1459 | 1559 | 1850 | > | (2029 x 4) |
L | L | L | L | Sum | |||||||
3 | 14 | 16 | 35 | 41 | 61 | 80 | 86 | 102 | > | 438 | |
AP-b | 1974 | 1739 | 1704 | 1377 | 1258 | 874 | 570 | 470 | 179 | > | (2029 x 5) |
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 23
ISSN 2229-5518
7 | 10 | 11 | 14 | 30 | 77 | 82 | 102 | 105 | > | 438 | |
AP-a | 150 | 211 | 233 | 290 | 572 | 1400 | 1494 | 1850 | 1916 | > | (2029 x 4) |
L | L | L | Sum | ||||||||
7 | 10 | 11 | 14 | 30 | 77 | 82 | 102 | 105 | > | 438 | |
AP-b | 1879 | 1818 | 1796 | 1739 | 1457 | 629 | 535 | 179 | 113 | > | (2029 x 5) |
etc.
The result of the research that we have carried out clearly shows that there is a matrix code in insulin. It also shows that the coding system within the amino acidic language gives a full information, not only for the amino acid „record“, but also for its structure, configuration and its various shapes. In the following text we shall discuss the issue of the existence of the insulin code, and also the issue of coding of individual structural levels in this protein.
A similar balance is established among all the other amino acids. Although those amino acids have different number of atoms, these numbers, when put into correlation with codes 19 and 7, give the same mathematical result, which is discrete code 2029. This goes for all the sequences in biochemistry. All the sequences, those with identical as well as those with different numerical values, when put into correlation with codes 19 and 7, give one result only. In this way, a global mathematical balance is established among sequences in nature. This means that the mathematical balance can be established even when sequences are not in balance.
In the previous examples we translated the physical and chemical parameters from the language of biochemistry into the digital language of programmatic, cybernetic and information principles. This we did by using the adequate mathematical algorithms. By using chemical- information procedures, we calculated the numerical value for the information content of molecules. What we got this way is the digital picture of the phenomenon of biochemistry. These digital pictures reveal to us a whole new dimension of this science. They reveal to us that the biochemical process is strictly conditioned and determined by programmatic, cybernetic and information principles.
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 24
ISSN 2229-5518
From the previous examples we can see that this protein really has its quantitative characteristics. It can be concluded that there is a connection between quantitative characteristics in the process of transfer of genetic information and the qualitative appearance of given genetic processes.
The results of our research show that the processes of sequencing the molecules are conditioned and arranged not only with chemical and biochemical lawfulness, but also with program, cybernetic and informational lawfulness too. At the first stage of our research we replaced nucleotides from the Amino Acid Code Matrix with numbers of the atoms and atomic numbers in those nucleotides. Translation of the biochemical language of these amino acids into a digital language may be very useful for developing new methods of predicting protein sub- cellular localization, membrane protein type, protein structure secondary prediction or any other protein attributes.
The success of human genome project has generated deluge of sequence information. The explosion of biological data has challenged scientists to accelerate the speed for their analysis. Nowadays, protein sequences are generally stored in the computer database system in the form of long character strings. It would act like a snail's pace for human beings to read these sequences with the naked eyes (Xiao and Chou, 2007). Also, it is very hard to extract any key features by directly reading these long character strings. However, if they can be converted to some signal process, many important features can be automatically manifested and easily studied by means of the existing tools of information theory (Xiao and Chou,
2007). The novel approach as presented here may help improve this kind of situation.
The process of sequencing in bio-macromolecules is conditioned and determined not only through biochemical, but also through cybernetic and information principles. The digital pictures of biochemistry provide us with cybernetic and information interpretation of the scientific facts. Now we have the exact scientific proofs that there is a genetic language that can be described by the theory of systems and cybernetics, and which functions in accordance with certain principles.
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 25
ISSN 2229-5518
[1] Cai, Y.D., and Chou, K.C., 2006. Predicting membrane protein type by functional
Domain composition and pseudo amino acid composition. J Theor Biol 238, 395-400. [2] Cai, Y.D., Zhou, G.P., and Chou, K.C., 2003. Support vector machines for predicting
membrane protein types by using functional domain composition. Biophys J 84, 3257-
3263.
[3] Cai, Y.D., Zhou, G.P., and Chou, K.C., 2005. Predicting enzyme family classes by hybridizing gene product composition and pseudo-amino acid composition. J Theor Biol
234, 145-149.
[4] Cai, Y.D., Feng, K.Y., Lu, W.C., and Chou, K.C., 2006. Using LogitBoost classifier to predict protein structural classes. J Theor Biol 238, 172-176.
[5] Cai, Y.D., Pong-Wong, R., Feng, K., Jen, J.C.H., and Chou, K.C., 2004. Application of
SVM to predict membrane protein types. J Theor Biol 226, 373-376.
[6] Chen, C., Chen, L., Zou, X., and Cai, P., 2009. Prediction of protein secondary structure content by using the concept of Chou's pseudo amino acid composition and support vector machine. Protein & Peptide Letters 16, 27-31.
[7] Chen, L., Feng, K.Y., Cai, Y.D., Chou, K.C., and Li, H.P., 2010. Predicting the network of substrate-enzyme-product triads by combining compound similarity and functional domain composition. BMC Bioinformatics 11, 293.
[8] Chou, K.C., 2011. Some remarks on protein attribute prediction and pseudo amino acid composition (50th Anniversary Year Review). J Theor Biol 273, 236-247.
[9] Chou, K.C., and Shen, H.B., 2010a. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Natural Science
2, 1090-1103 (openly accessible at http://www.scirp.org/journal/NS/).
[10] Chou, K.C., and Shen, H.B., 2010b. Plant-mPLoc: A Top-Down Strategy to Augment the
Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 5, e11335.
[11] Ding, H., Luo, L., and Lin, H., 2009a. Prediction of cell wall lytic enzymes using Chou's amphiphilic pseudo amino acid composition. Protein & Peptide Letters 16, 351-355.
[12] Ding, Y.S., Zhang, T.L., and Chou, K.C., 2007. Prediction of protein structure classes with pseudo amino acid composition and fuzzy support vector machine network. Protein
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 26
ISSN 2229-5518
& Peptide Letters 14, 811-815.
[13] Ding, Y.S., Zhang, T.L., Gu, Q., Zhao, P.Y., and Chou, K.C., 2009b. Using maximum entropy model to predict protein secondary structure with single sequence. Protein & Peptide Letters 16, 552-560.
[14] He, Z.S., Zhang, J., Shi, X.H., Hu, L.L., Kong, X.G., Cai, Y.D., and Chou, K.C., 2010.
Predicting drug-target interaction networks based on functional groups and biological features. PLoS ONE 5, e9603.
[15] Hu, L., Huang, T., Shi, X., Lu, W.C., Cai, Y.D., and Chou, K.C., 2011. Predicting functions of proteins in mouse based on weighted protein-protein interaction network and protein hybrid properties PLoS ONE 6, e14556.
[16] Huang, T., Shi, X.H., Wang, P., He, Z., Feng, K.Y., Hu, L., Kong, X., Li, Y.X., Cai,
Y.D., and Chou, K.C., 2010. Analysis and prediction of the metabolic stability of proteins based on their sequential features, subcellular locations and interaction networks PLoS ONE 5, e10972.
[17] Kandaswamy, K.K., Pugalenthi, G., Moller, S., Hartmann, E., Kalies, K.U., Suganthan, P.N., and Martinetz, T., 2010. Prediction of Apoptosis Protein Locations with Genetic Algorithms and Support Vector Machines Through a New Mode of Pseudo Amino Acid Composition. Protein and Peptide Letters 17, 1473-1479.
[18] Kannan, S., Hauth, A.M., and Burger, G., 2008. Function prediction of hypothetical proteins without sequence similarity to proteins of known function. Protein & Peptide Letters 15, 1107-1116.
[19] Liu, T., Zheng, X., Wang, C., and Wang, J., 2010. Prediction of Subcellular Location of
Apoptosis Proteins using Pseudo Amino Acid Composition: An Approach from Auto
Covariance Transformation. Protein & Peptide Letters 17, 1263-9.
[20] Mohabatkar, H., 2010. Prediction of cyclin proteins using Chou's pseudo amino acid composition. Protein & Peptide Letters 17, 1207-1214.
[21] Wang, Y.C., Wang, X.B., Yang, Z.X., and Deng, N.Y., 2010. Prediction of enzyme subfamily class via pseudo amino acid composition by incorporating the conjoint triad feature. Protein & Peptide Letters 17, 1441-1449.
[22] Xiao, X., and Chou, K.C., 2007. Digital coding of amino acids based on hydrophobic index. Protein & Peptide Letters 14, 871-875.
IJSER © 2011 http://www.ijser.org
International Journal of Scientific & Engineering Research Volume 2, Issue 8, August-2011 27
ISSN 2229-5518
IJSER <S>2011
http:1/Y'Nffl .ijser .ora