International Journal of Scientific & Engineering Research Volume 2, Issue 12, December-2011 1
ISSN 2229-5518
A Novel Hybrid Fuzzy Clustering based approach for the effective Quantification and Analysis of cDNA Microarray Images.
A.Sri Nagesh, Dr.G.P.Saradhi Varma, Dr A Govardhan
Abstract — In this paper, we propose a hybrid approach for microarray image analysis, which is to quantify the intensity of each spot and lo- cate differentially articulated genes with the aid of image processing and machine learning techniques. Initially we employ a hill-climbing au- tomatic gridding and spot quantification technique, which takes a microarray image (or a sub-grid) as input, and makes no assumptions con- cerning the size of the spots, rows and columns in the grid. We propose an approach based on image processing techniques for microarray image segmentation that includes a noise-removal pre-processing stage. The foreground and background pixels from the microarray images are segmented with the aid of morphological operator and common subtraction procedure whereas the noise is filtered by using wiener fil- tering. Finally for cluster analysis we employed a hybrid approach based on clustering techniques; Fuzzy C Means and Fuzzy K Means. Cluster- ing and their analysis were performed on this inputted microarray data. To quantify the effectiveness of the proposed approach, we utilized the Microarray database which is available publicly and we evaluated the accuracy, the specificity and the sensitivity of our proposed ap- proach.
Index Terms — Bioinformatics, DNA Microarray Gene Expression, Gridding, Hill Climbing, Image Segmentation, Morphological Operators, Hybrid clustering. Microarray Analysis Normalization, Spot Localization, Wiener Filter,
—————————— ——————————
N this paper we propose a hybrid approach for microarray image analysis, which is to quantify the intensity of each spot and locate differentially expressed genes with the aid
of image processing and machine learning techniques. The analysis of the images, as seen is not a trivial task – it involves gridding, segmentation, normalization, quantification, statis- tical and cluster analysis. Of these, we worked in some of the areas such as DNA microarray image gridding, segmentation and cluster analysis. Gridding is necessary to accurately iden- tify the location of each spot while extracting spot intensities from the microarray images. For gridding we devised an ap- proach based on hill-climbing, which is competent to locate the grid with high accuracy on standard dataset images and exploiting a least number of parameters. Next we aspired to deal with the problem of microarray image segmentation. In micro array, segmentation refers to the classification of pixels as either foreground (represent the signal) or background (represent the surrounding area). We proposed an approach for microarray image segmentation based on image processing
————————————————
A.Sri Nagesh is currently doing Ph.D in Image Processing area in JNTUni- versity, Hyderabad, India, E-mail: asringesh@gmail.com.
Dr.G.P.Saradhi Varma is working as Professor , & HOD, IT Department
SRKR Engineering College, Bhimavaram, India. E-mail:gpsvarma@yahoo.com
Dr.A.Govardhan is working as Principal & Professor, CSE Deparment,
JNTUH, Jagityal, India,. E-mail: govardhan_cse@yahoo.co.in
techniques that includes a noise-removal stage. The fore- ground and background pixels from the microarray image are segmented with the aid of common subtraction procedure and the noise is filtered by using wiener filter.
Finally for cluster analysis we proposed a hybrid approach based on clustering techniques such as Fuzzy C-means and Fuzzy K-means clustering. Clustering is the grouping of the objects that are more similar to each other. We examine the application of hybrid clustering to microarray data analysis, and then we compare the performance of this hybrid cluster- ing method with the existing clustering methods. We also eva- luate each of this clustering method with validation measures for real-life datasets.The rest of the paper is organized as fol- lows Section 2 presents a brief review of some recent signifi- cant researches in Microarray image analysis. The proposed approach for segmentation of microarray image is explained in the paper is given in section 3 where a detailed the pro- posed methodology for cluster analysis. Experimental results and analysis of the proposed methodology are discussed in Section 4. Finally, concluding remarks are provided in Section
5.
IJSER © 2011
International Journal of Scientific & Engineering Research Volume 2, Issue 12, December-2011 2
ISSN 2229-5518
In this section, a brief review of some important contributions from the existing literature is presented.
Wu, H. and Yan, H. [1] have addressed the segmentation and information extraction problems. They have implemented a segmentation method based on K-means clustering, a back- ground and foreground correction algorithm based on ma- thematical morphological and histogram analysis for informa- tion extraction. It does not have any restrictions for the shape of spots and this served as an advantage to their method. Ex- perimental results are being compared with those attained from the genuine software GenePix.
A method is being portrayed by Nikolaos Giannakeas and Dimitrios I. Fotiadis [2] for the automated analysis of microar- ray images. Gridding and Segmentation are the two stages of their proposed method. Template matching is initially used to preprocess the microarray images; afterwards, block and spot finding process are being done. Afterwards, the non-expressed spots are being identified and using a Voronoi diagram, a grid is fit on the image. K-means and Fuzzy C means (FCM) clus- tering are employed in the segmentation stage. Images from the Stanford Microarray Database (SMD) are used to evaluate their proposed method. Contrasting with the two previously developed K-means-based methods, the efficiency of our Fuzzy C means-based work is shown in the results presented at the segmentation stage. Images with artifacts can be easily handled by their proposed method and it is entirely robotic.
1. Assign every gene to its own cluster.
2. Trace the nearby pair of clusters and merge them into
a single cluster.
3. Calculate the distances (similarities) between the new cluster and each of the old clusters with the distance
measure method.
4. Reiterate steps 2 and 3 till all genes are clustered.
A variety of experiments have been performed to eva- luate the proposed methodology for the analysis of DNA mi- croarray images. The artificially constructed or the publicly available database that belongs to Lymphoma/Leukemia Mo- lecular Profiling Project Gateway are taken and utilized as the test images [4]. The goal of this study is to experiment and compare the approaches for the microarray image analysis process. In our experiments, we apply hill climbing to perform Gridding, the foreground and background pixels are seg- mented with the aid of common background subtraction pro- cedure and finally we employ hybrid clustering approach for cluster analysis. The proposed researches have been imple- mented in Matlab (Matlab version 7.10). We have conducted experiments to examine the performance reliability of the fuzzy-type clustering methods. The resulted images are de- picted in figure 1. The accuracy, specificity and sensitivity of proposed hybrid clustering based methodology in comparison with fuzzy c means and fuzzy k means clustering techniques are depicted in table. 1. Whereas the accuracy, the specificity and the sensitivity of the proposed method is termed as:
Accuracy
![]()
No. of
correctly
det ected
pixels
Cluster analysis is the process of grouping (clustering) large data sets based on the similarity criteria for appropriate- ly scaled variables that represent the data of interest. Genes or
Total
No. of
pixels
in the
image
(1)
samples are grouped into "clusters" on the basis of the similar
expression profiles in Cluster analysis and bestows clues to
Specificit y
![]()
No. of
correctly
identified signal
pixels
the function or regulation of genes or similarity of samples by means of shared cluster membership. To analyze genome- wide expression data, numerous clustering models have been practiced. In our work, we apply a hybrid approach based on
Total
No. of
signal pixels
(2)
clustering techniques for cluster analysis. The clustering tech-![]()
Sensitivity No. of
correctly
identified background
pixels
niques utilized in the proposed approach are a combination of
fuzzy C means and fuzzy k means clustering.
The hybrid clustering approach with the combination of Fuzzy C means and Fuzzy K means clustering is done
based on the following step for a set of N genes to be clus-
tered, and a NxN distance (or similarity) matrix,
Total
No. of
background pixels
(3)
IJSER © 2011
International Journal of Scientific & Engineering Research Volume 2, Issue 12, December-2011 3


ISSN 2229-5518

(a) (b)
(c)
(d)
(e)
Figure 1: Spotted microarray image (a) Input Image, (b) Grayscale image, (c) Gridded image, (d) Binary image, (e) Filtered image.
As a simple baseline for comparison, nearest neighbor classifi- er with Euclidean distance is used. The nearest neighbor clas- sifier simply classifies a test instance with the class of the nearest training instance according to some distance measure. Performance of each parameter is measured by comparing the spot detection results with an expert’s ground truth. Three performance measurements, namely, true positive (TP, a number of foreground spot pixels correctly detected), false positive (FP, a number of spot pixels which are detected wrongly as spot intensity pixels), false negative (FN, a number of foreground spot pixels that are not detected), true negative (TN, a number of background spot pixels which are correctly identified as non spot pixels), sensitivity, specificity, show the computation of sensitivity, specificity, and accuracy, respec- tively:
Accuracy = TP+ TN/TP+FP+ FN+TN (4) Specificity = TN /TN +FP (5) Sensitivity = TP/TP+ FN (6)
Table 1: Comparison of Accuracy, specificity and sensitivity results for the proposed hybrid approach and other clustering based methods.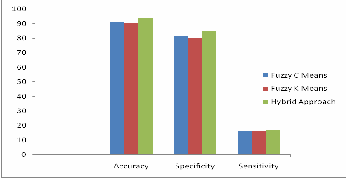
IJSER © 2011
International Journal of Scientific & Engineering Research Volume 2, Issue 12, December-2011 4
ISSN 2229-5518
Figure 2: Comparison Graph showing all the three techniques.
DNA Microarrays are powerful techniques that are utilized to analyze the expression of DNA in organisms sub- sequent to performing experiments. In this paper, a hybrid clustering-based approach for the analysis of microarray im- ages has been presented. The proposed research is a multi- channel approach consisting of three steps, the gridding step where hill climbing approach is applied to the initial image to identify the location of the spots, then the segmentation step where all the pixels of the image are classified into signal, background or artifacts by using morphological operator and common background subtraction procedure, whereas the arti- facts are removed by employing wiener filter. Finally cluster analysis is carried out with the aid of hybrid clustering ap- proach. The clustering techniques employed are Fuzzy C means and Fuzzy K means. The experimental results have illustrated the effectiveness of the proposed methodology for cluster analysis. Thus the effectiveness of our algorithm has been tested on datasets drawn from standard experiments, showing that our approach can effectively cluster the datasets based on profile similarity. According to the very promising accuracy results, the precision of the microarray data during experimentation might be significantly influenced.
The authors would like to thank all the authors and people who are directly or indirectly contributed for the outcome of this paper.
[1] Wu, H., Yan, H., ―Microarray Image Processing Based on Clustering and Morphological Analysis‖, In First Asia Pacific Bioinformatics Conference, 111-118, 2003.
[2] N. Giannakeas, D. Fotiadis, "An automated method for gridding and
clustering-based segmentation of cDNA microarray images", Com- puterized Medical Imaging and Graphics, Vol. 33, No. 1, pp. 40-49,
2009.
[3] A.Sri Nagesh, Dr.A.Govardhan, Dr G.P.S.Varma, Dr G.S.Prasad,‖An Automated Histogram Equalized Fuzzy Clustering based Approach for the Segmentation of Microarray images.‖ ANU Journal of Engi- neering and Technology, pp 42-48., Vol 2, Issue 2, December 2010. ISSN: 0976-3414.
[4] “Microarray Images”,from http://llmpp.nih.gov/lymphoma/data/rawdata/
[5] Yong Han, "Multi-Objective Differential Evolution for Automatic Clustering with Application to Micro-Array Data Analysis", Sensors, Vol. 9, pp. 3981-4004, 2009
[6] C. W. Whitfield, A. M. Cziko, and G. E. Robinson, ―Gene expression profiles in the brain predict behavior in individual honey bees,‖ Science, vol. 302, pp. 296–299, 2003.
[7] P. Bajcsy, ―Gridline: automatic grid alignment in DNA microarray scans,‖ IEEE Transactions on Image Processing, vol. 13, no. 1, pp. 15–
25, 2004.
[8] A. W.-C. Liew, H. Yang, and M. Yang, ―Robust adaptive spot seg-
mentation of DNA microarray images,‖ Pattern Recogn., vol. 36, pp.
1251–1254, 2003.
[9] M. Katzer, F. Kummert, and G. Sageter, ―AMarkov random field model of microarray gridding,‖ in Proc. ACMSymp. Applied Com- puting (SAC), Melbourne, FL, pp. 72–77, 2003.
[10] Rahnenführer, J., & Bozinov, V., ―Hybrid clustering for microarray image analysis combining intensity and shape features‖, BMC Bioin- formatics, Vol. 5, No. 47, 2004.
[11] B. J. Oommen and L. Rueda, ―A Formal Analysis of Why Heuristic
Functions Work", Artificial Intelligence, Vol.164, pp.1–22, 2005.
[12] V. Vidyadharan, "Automatic Gridding of DNA Microarray Images", Master’s thesis, School of Computer Science, University of Windsor, Canada, 2004.Electronically available at http://cs.uwindsor.ca/~lrueda/papers /VidyaThesis.pdf.
[13] Luis Rueda and Vidya Vidyadharan, "A Hill-climbing Approach for
Automatic Gridding of cDNA Microarray Images", IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB), Vol. 3, No. 1, pp. 72, January 2006.
[14] Iiris Hovatta, Katja Kimppa, Antti Lehmussola, Tomi Pasanen et al., "DNA Microarray Data Analysis", Eds: - Jarno Tuimala and M. Min- na Laine, second edition, CSC - Scientific Computing Ltd., Finland, pages: 165, 2005.
[15] Chinatsu Arima, Taizo Hanai, "Gene Expression Analysis Using
Fuzzy K-Means Clustering", Genome Informatics, Vol. 14, pp. 334-
335, 2003.
[16] ―Unsharp Filter‖ from http://homepages.inf.ed.ac.uk/rbf/HIPR2/
[17] J. C. Bezdek, "Pattern Recognition with Fuzzy Objective Function
Algorithms", Kluwer Academic Publishers, Norwell, MA, USA, 1981. [18] A.Sri Nagesh, Dr G.P.S.Varma, Dr.A.Govardhan ―An Improved Iter- ative Watershed and Morphological Transformation Techniques for Segmentation of Microarray Images‖ IJCA Special Issue on ―Com-
puter Aided Soft Computing Techniques for Imaging and Biomedical
Applications‖ CASCT, 2010.pp-77-87, ISSN: 0975-8887.
[19] Nagarajan, R., ―Intensity-Based Segmentation of Microarray Images‖,
IEEE Transactions on Medical Imaging, Vol. 22, No. 7, pp. 882–889,
2003.
[20] Kaushik Suresh, Debarati Kundu, Sayan Ghosh, Swagatam Das, Ajith Abraham and Sang Yong Han, "Multi-Objective Differential Evolu- tion for Automatic Clustering with Application to Micro-Array Data Analysis", Sensors, Vol. 9, pp. 3981-4004, 2009.
[21] Volkan Uslan and Dhsan Omur Bucak, "Microarray Image Segmenta- tion Using Clustering Methods", Mathematical and Computational Applications, Vol. 15, No. 2, pp. 240-247, 2010.
IJSER © 2011